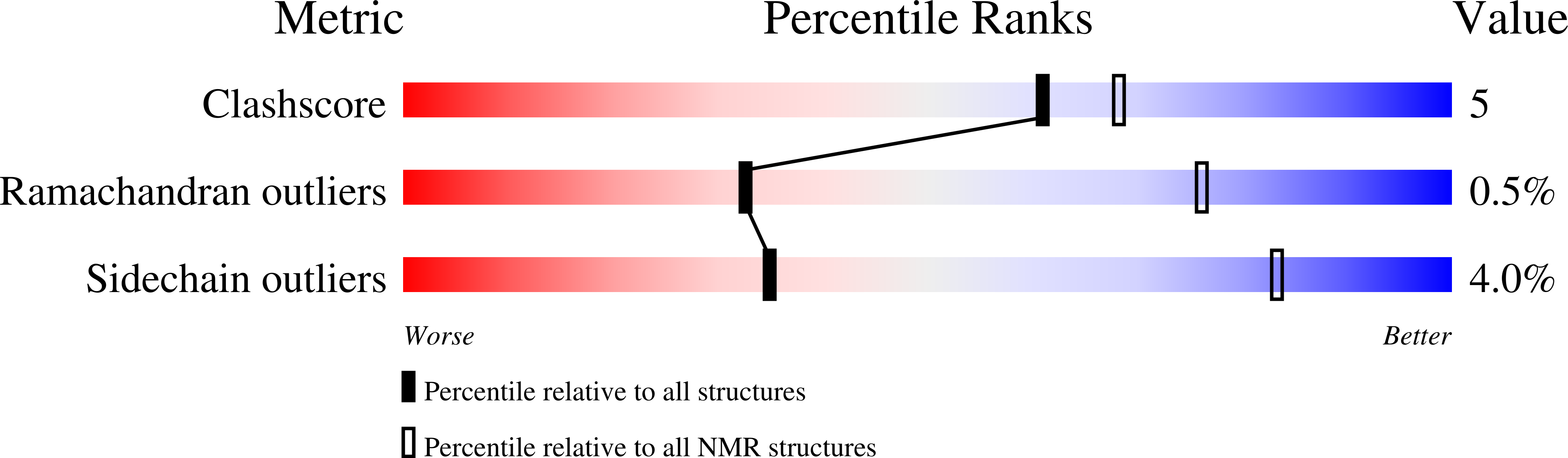Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
Weinheimer, A.S., Paung, Y., Rageul, J., Khan, A., Lo, N., Ho, B., Tong, M., Alphonse, S., Seeliger, M.A., Kim, H.(2022) J Biol Chem 298: 102268-102268
- PubMed: 35850305
- DOI: https://doi.org/10.1016/j.jbc.2022.102268
- Primary Citation of Related Structures:
7N99 - PubMed Abstract:
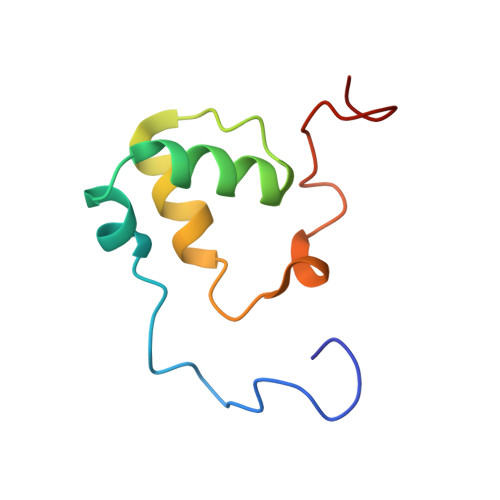
Elevated DNA replication stress causes instability of the DNA replication fork and increased DNA mutations, which underlies tumorigenesis. The DNA replication stress regulator silencing-defective 2 (SDE2) is known to bind to TIMELESS (TIM), a protein of the fork protection complex, and enhances its stability, thereby supporting replisome activity at DNA replication forks. However, the DNA-binding activity of SDE2 is not well defined. Here, we structurally and functionally characterize a new conserved DNA-binding motif related to the SAP (SAF-A/B, Acinus, PIAS) domain in human SDE2 and establish its preference for ssDNA. Our NMR solution structure of the SDE2 SAP domain reveals a helix-extended loop-helix core with the helices aligned parallel to each other, consistent with known canonical SAP folds. Notably, we have shown that the DNA interaction of this SAP domain extends beyond the core SAP domain and is augmented by two lysine residues in the C-terminal tail, which is uniquely positioned adjacent to the SAP motif and conserved in the pre-mRNA splicing factor SF3A3. Furthermore, we found that mutation in the SAP domain and extended C terminus not only disrupts ssDNA binding but also impairs TIM localization at replication forks, thus inhibiting efficient fork progression. Taken together, our results establish SDE2 SAP as an essential element for SDE2 to exert its role in preserving replication fork integrity via fork protection complex regulation and highlight the structural diversity of the DNA-protein interactions achieved by a specialized DNA-binding motif.
Organizational Affiliation:
Department of Biochemistry and Cell Biology, State University of New York at Stony Brook, Stony Brook, New York, USA.