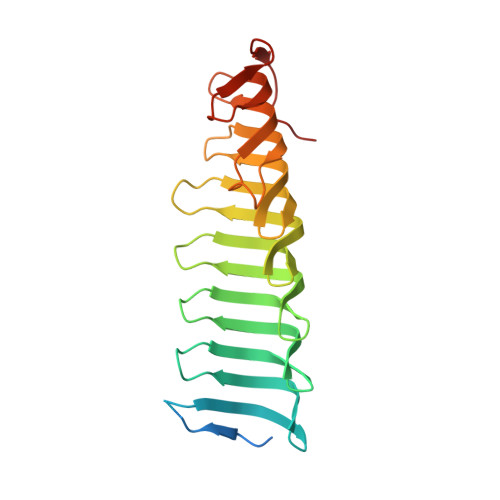
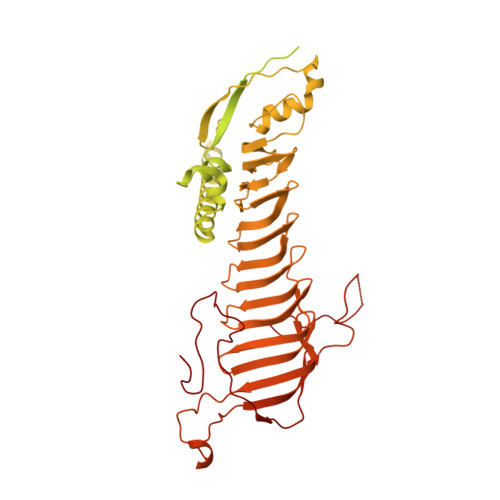
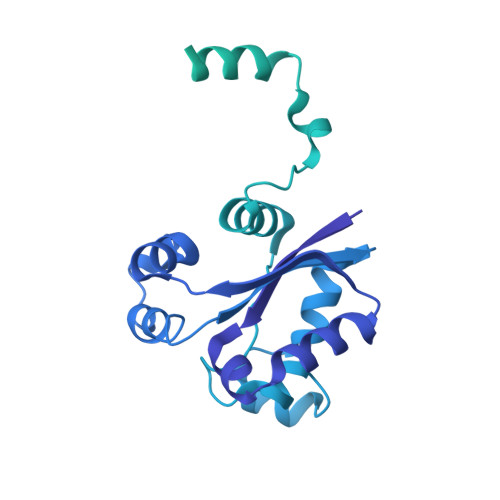
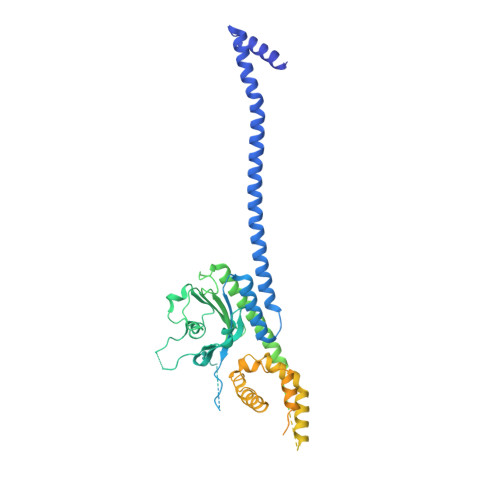
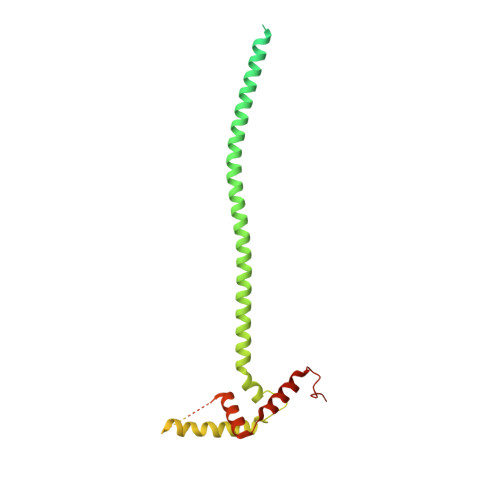
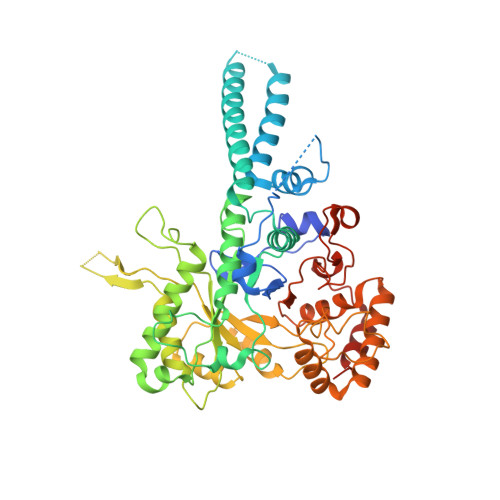
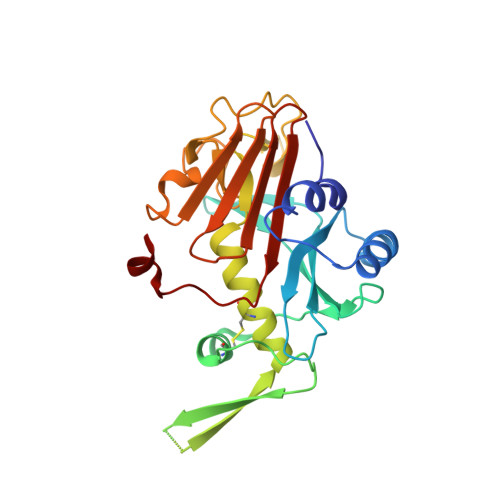
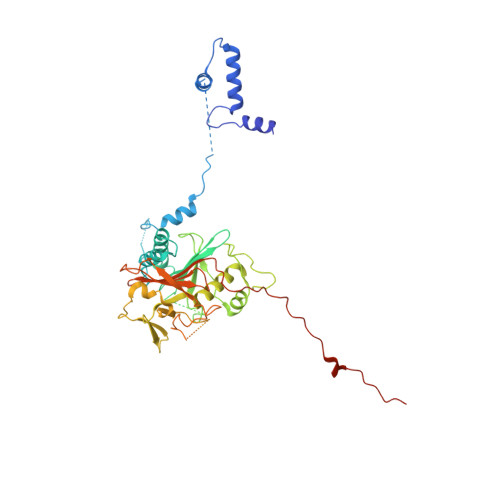
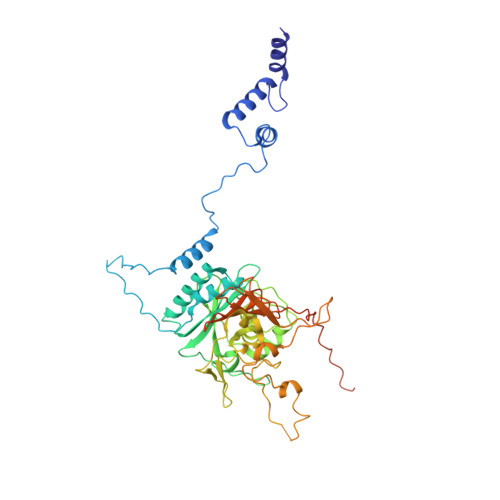
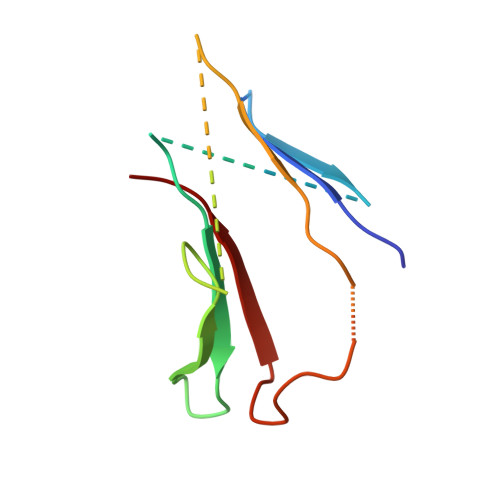
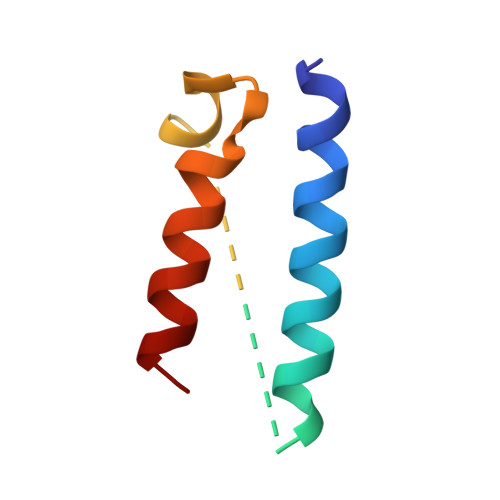
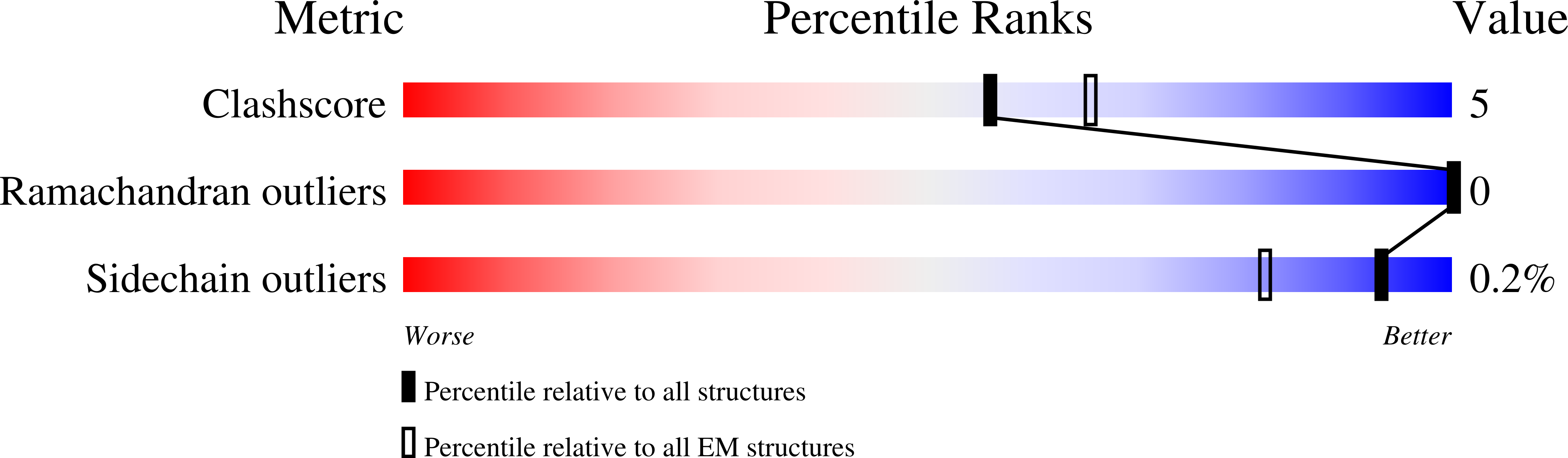Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Grossman-Haham, I., Coudray, N., Yu, Z., Wang, F., Zhang, N., Bhabha, G., Vale, R.D.(2021) Nat Struct Mol Biol 28: 20-28
- PubMed: 33318704
- DOI: https://doi.org/10.1038/s41594-020-00519-9
- Primary Citation of Related Structures:
7JR9, 7JRJ - PubMed Abstract:
Motile cilia power cell locomotion and drive extracellular fluid flow by propagating bending waves from their base to tip. The coordinated bending of cilia requires mechanoregulation by the radial spoke (RS) protein complexes and the microtubule central pair (CP). Despite their importance for ciliary motility across eukaryotes, the molecular function of the RSs is unknown. Here, we reconstituted the Chlamydomonas reinhardtii RS head that abuts the CP and determined its structure using single-particle cryo-EM to 3.1-Å resolution, revealing a flat, negatively charged surface supported by a rigid core of tightly intertwined proteins. Mutations in this core, corresponding to those involved in human ciliopathies, compromised the stability of the recombinant complex, providing a molecular basis for disease. Partially reversing the negative charge on the RS surface impaired motility in C. reinhardtii. We propose that the RS-head architecture is well-suited for mechanoregulation of ciliary beating through physical collisions with the CP.
Organizational Affiliation:
Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, CA, USA.