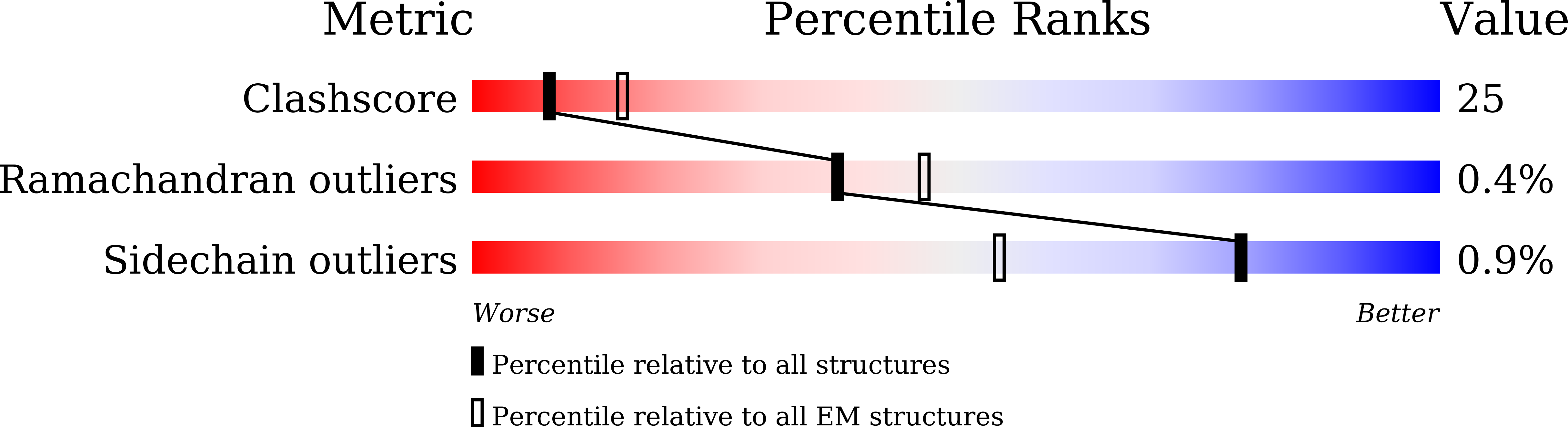The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors.
Zhu, G., Zeng, H., Zhang, S., Juli, J., Tai, L., Zhang, D., Pang, X., Zhang, Y., Lam, S.M., Zhu, Y., Peng, G., Michel, H., Sun, F.(2021) Angew Chem Int Ed Engl 60: 13323-13330
- PubMed: 33665933
- DOI: https://doi.org/10.1002/anie.202016785
- Primary Citation of Related Structures:
7DEG - PubMed Abstract:
The heme-copper oxidase superfamily comprises cytochrome c and ubiquinol oxidases. These enzymes catalyze the transfer of electrons from different electron donors onto molecular oxygen. A B-family cytochrome c oxidase from the hyperthermophilic bacterium Aquifex aeolicus was discovered previously to be able to use both cytochrome c and naphthoquinol as electron donors. Its molecular mechanism as well as the evolutionary significance are yet unknown. Here we solved its 3.4 Å resolution electron cryo-microscopic structure and discovered a novel dimeric structure mediated by subunit I (CoxA2) that would be essential for naphthoquinol binding and oxidation. The unique structural features in both proton and oxygen pathways suggest an evolutionary adaptation of this oxidase to its hyperthermophilic environment. Our results add a new conceptual understanding of structural variation of cytochrome c oxidases in different species.
Organizational Affiliation:
National Key Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing, 100101, China.