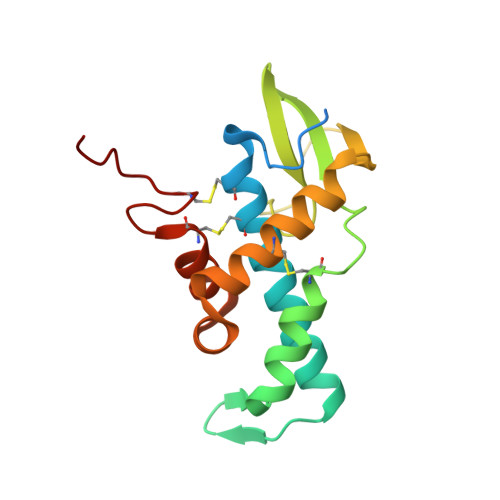
High-resolution Crystal Structure of Human pERp1, A Saposin-like Protein Involved in IgA, IgM and Integrin Maturation in the Endoplasmic Reticulum.
Sowa, S.T., Moilanen, A., Biterova, E., Saaranen, M.J., Lehtio, L., Ruddock, L.W.(2021) J Mol Biol 433: 166826-166826
- PubMed: 33453188
- DOI: https://doi.org/10.1016/j.jmb.2021.166826
- Primary Citation of Related Structures:
7AAH - PubMed Abstract:
The folding of disulfide bond containing proteins in the endoplasmic reticulum (ER) is a complex process that requires protein folding factors, some of which are protein-specific. The ER resident saposin-like protein pERp1 (MZB1, CNPY5) is crucial for the correct folding of IgA, IgM and integrins. pERp1 also plays a role in ER calcium homeostasis and plasma cell mobility. As an important factor for proper IgM maturation and hence immune function, pERp1 is upregulated in many auto-immune diseases. This makes it a potential therapeutic target. pERp1 belongs to the CNPY family of ER resident saposin-like proteins. To date, five of these proteins have been identified. All are implicated in protein folding and all contain a saposin-like domain. All previously structurally characterized saposins are involved in lipid binding. However, there are no reports of CNPY family members interacting with lipids, suggesting a novel function for the saposin fold. However, the molecular mechanisms of their function remain elusive. To date, no structure of any CNPY protein has been reported. Here, we present the high-resolution (1.4 Å) crystal structure of human pERp1 and confirm that it has a saposin-fold with unique structural elements not present in other saposin-fold structures. The implications for the role of CNPY proteins in protein folding in the ER are discussed.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine, University of Oulu, Aapistie 7, 90220 Oulu, Finland; Biocenter Oulu, Aapistie 5, 90220 Oulu, Finland. Electronic address: sven.sowa@oulu.fi.