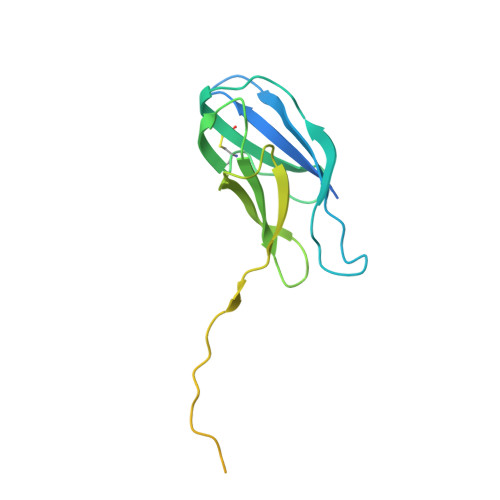
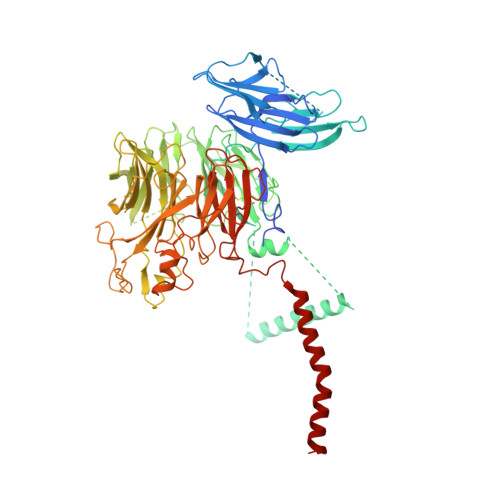
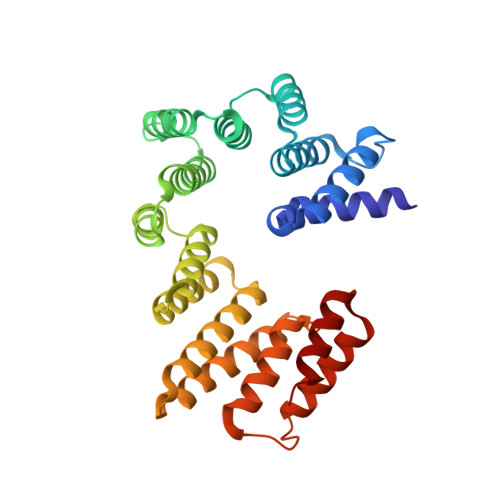
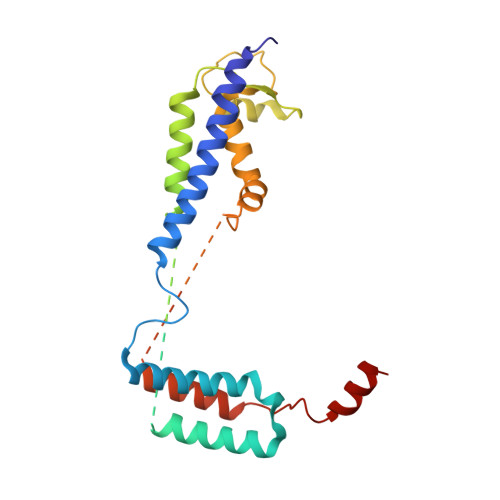
Structure of the ER membrane complex, a transmembrane-domain insertase.
Bai, L., You, Q., Feng, X., Kovach, A., Li, H.(2020) Nature 584: 475-478
- PubMed: 32494008
- DOI: https://doi.org/10.1038/s41586-020-2389-3
- Primary Citation of Related Structures:
6WB9 - PubMed Abstract:
The endoplasmic reticulum (ER) membrane complex (EMC) cooperates with the Sec61 translocon to co-translationally insert a transmembrane helix (TMH) of many multi-pass integral membrane proteins into the ER membrane, and it is also responsible for inserting the TMH of some tail-anchored proteins 1-3 . How EMC accomplishes this feat has been unclear. Here we report the first, to our knowledge, cryo-electron microscopy structure of the eukaryotic EMC. We found that the Saccharomyces cerevisiae EMC contains eight subunits (Emc1-6, Emc7 and Emc10), has a large lumenal region and a smaller cytosolic region, and has a transmembrane region formed by Emc4, Emc5 and Emc6 plus the transmembrane domains of Emc1 and Emc3. We identified a five-TMH fold centred around Emc3 that resembles the prokaryotic YidC insertase and that delineates a largely hydrophilic client protein pocket. The transmembrane domain of Emc4 tilts away from the main transmembrane region of EMC and is partially mobile. Mutational studies demonstrated that the flexibility of Emc4 and the hydrophilicity of the client pocket are required for EMC function. The EMC structure reveals notable evolutionary conservation with the prokaryotic insertases 4,5 , suggests that eukaryotic TMH insertion involves a similar mechanism, and provides a framework for detailed understanding of membrane insertion for numerous eukaryotic integral membrane proteins and tail-anchored proteins.
Organizational Affiliation:
Structural Biology Program, Van Andel Institute, Grand Rapids, MI, USA. lin.bai@vai.org.