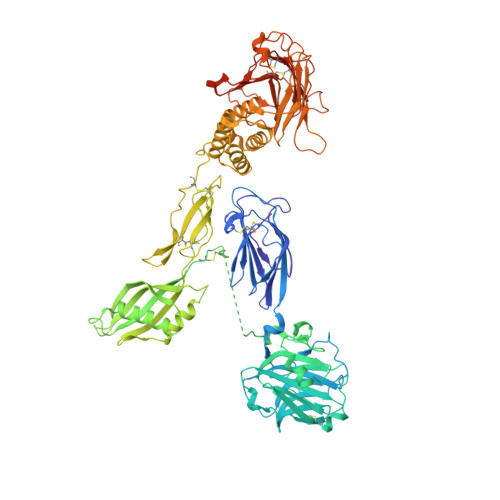
Structural basis for adhesion G protein-coupled receptor Gpr126 function.
Leon, K., Cunningham, R.L., Riback, J.A., Feldman, E., Li, J., Sosnick, T.R., Zhao, M., Monk, K.R., Arac, D.(2020) Nat Commun 11: 194-194
- PubMed: 31924782
- DOI: https://doi.org/10.1038/s41467-019-14040-1
- Primary Citation of Related Structures:
6V55 - PubMed Abstract:
Many drugs target the extracellular regions (ECRs) of cell-surface receptors. The large and alternatively-spliced ECRs of adhesion G protein-coupled receptors (aGPCRs) have key functions in diverse biological processes including neurodevelopment, embryogenesis, and tumorigenesis. However, their structures and mechanisms of action remain unclear, hampering drug development. The aGPCR Gpr126/Adgrg6 regulates Schwann cell myelination, ear canal formation, and heart development; and GPR126 mutations cause myelination defects in human. Here, we determine the structure of the complete zebrafish Gpr126 ECR and reveal five domains including a previously unknown domain. Strikingly, the Gpr126 ECR adopts a closed conformation that is stabilized by an alternatively spliced linker and a conserved calcium-binding site. Alternative splicing regulates ECR conformation and receptor signaling, while mutagenesis of the calcium-binding site abolishes Gpr126 function in vivo. These results demonstrate that Gpr126 ECR utilizes a multi-faceted dynamic approach to regulate receptor function and provide relevant insights for ECR-targeted drug design.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, 60637, USA.