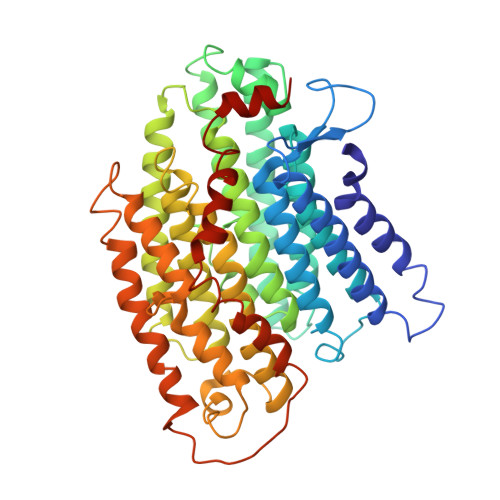
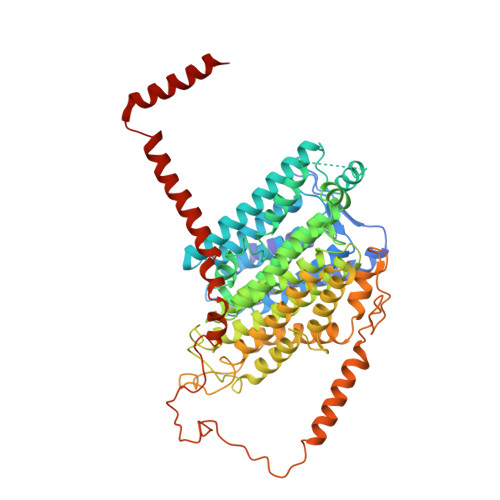
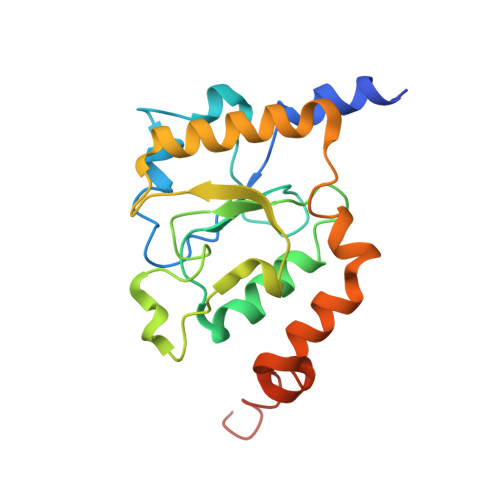
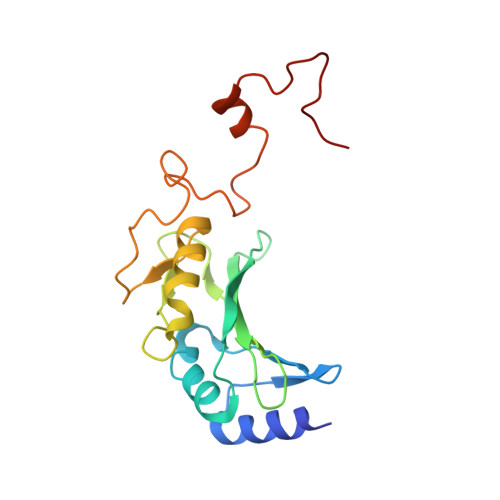
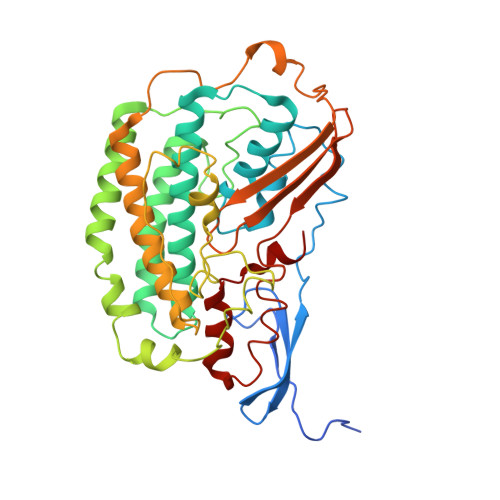
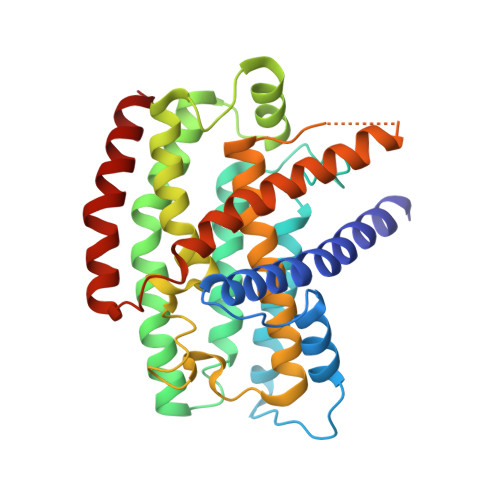
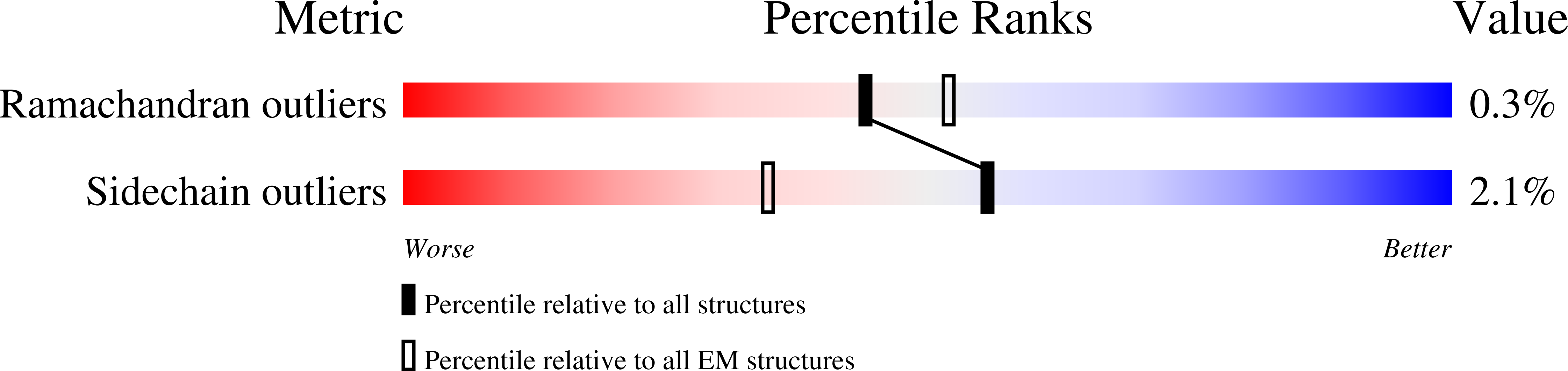Structure of the respiratory MBS complex reveals iron-sulfur cluster catalyzed sulfane sulfur reduction in ancient life.
Yu, H., Haja, D.K., Schut, G.J., Wu, C.H., Meng, X., Zhao, G., Li, H., Adams, M.W.W.(2020) Nat Commun 11: 5953-5953
- PubMed: 33230146
- DOI: https://doi.org/10.1038/s41467-020-19697-7
- Primary Citation of Related Structures:
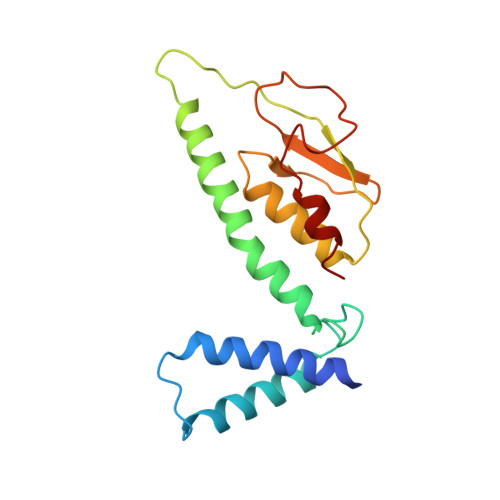
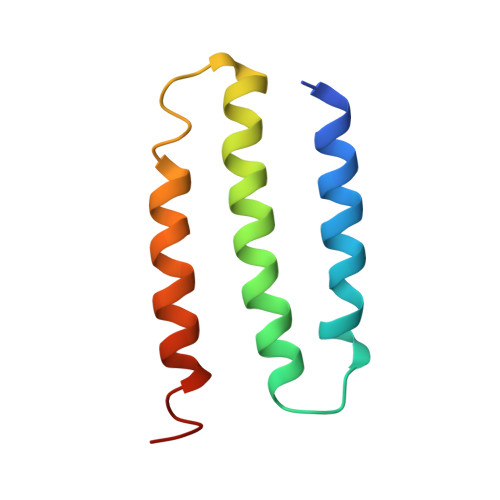
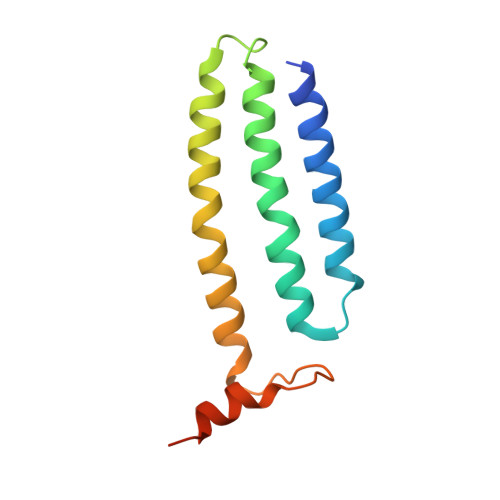
6U8Y - PubMed Abstract:
Modern day aerobic respiration in mitochondria involving complex I converts redox energy into chemical energy and likely evolved from a simple anaerobic system now represented by hydrogen gas-evolving hydrogenase (MBH) where protons are the terminal electron acceptor. Here we present the cryo-EM structure of an early ancestor in the evolution of complex I, the elemental sulfur (S 0 )-reducing reductase MBS. Three highly conserved protein loops linking cytoplasmic and membrane domains enable scalable energy conversion in all three complexes. MBS contains two proton pumps compared to one in MBH and likely conserves twice the energy. The structure also reveals evolutionary adaptations of MBH that enabled S 0 reduction by MBS catalyzed by a site-differentiated iron-sulfur cluster without participation of protons or amino acid residues. This is the simplest mechanism proposed for reduction of inorganic or organic disulfides. It is of fundamental significance in the iron and sulfur-rich volcanic environments of early earth and possibly the origin of life. MBS provides a new perspective on the evolution of modern-day respiratory complexes and of catalysis by biological iron-sulfur clusters.
Organizational Affiliation:
Structural Biology Program, Van Andel Institute, Grand Rapids, MI, USA.