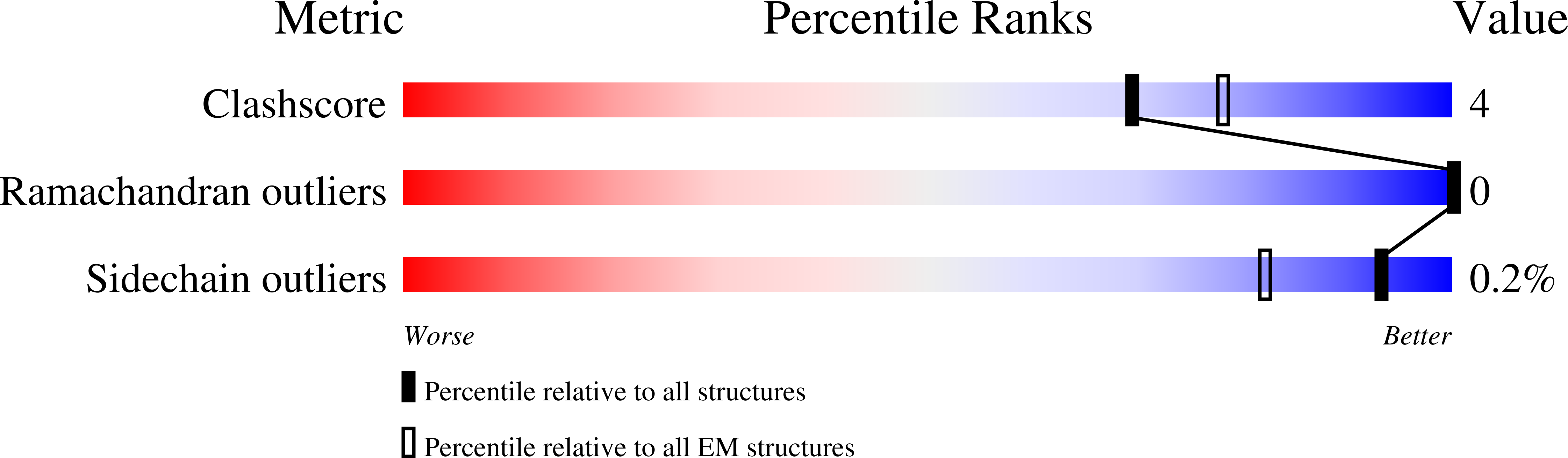ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Muhleip, A., Kock Flygaard, R., Ovciarikova, J., Lacombe, A., Fernandes, P., Sheiner, L., Amunts, A.(2021) Nat Commun 12: 120-120
- PubMed: 33402698
- DOI: https://doi.org/10.1038/s41467-020-20381-z
- Primary Citation of Related Structures:
6TMG, 6TMH, 6TMI, 6TMJ, 6TMK, 6TML - PubMed Abstract:
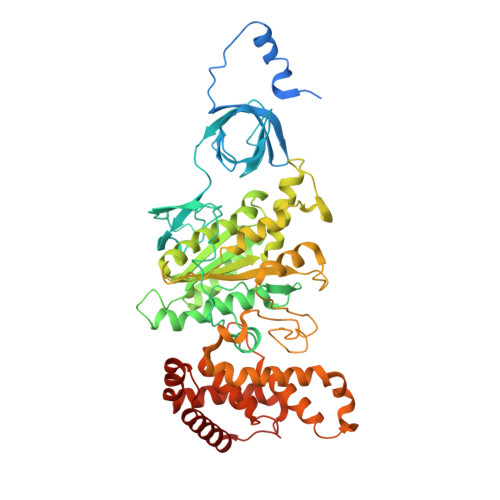
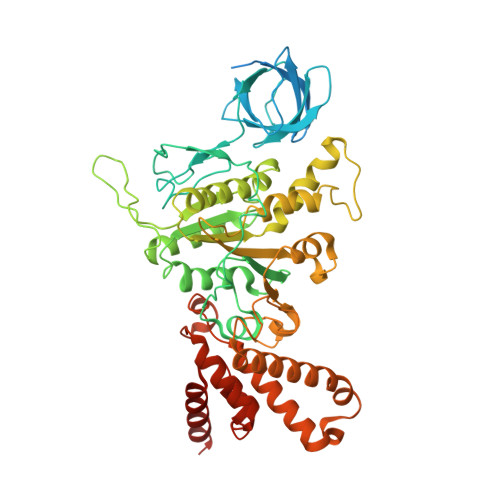
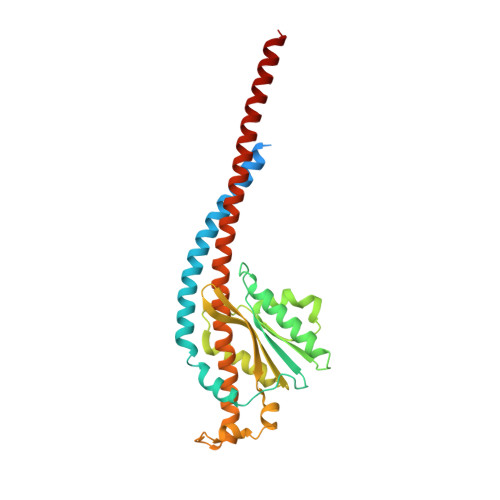
Mitochondrial ATP synthase plays a key role in inducing membrane curvature to establish cristae. In Apicomplexa causing diseases such as malaria and toxoplasmosis, an unusual cristae morphology has been observed, but its structural basis is unknown. Here, we report that the apicomplexan ATP synthase assembles into cyclic hexamers, essential to shape their distinct cristae. Cryo-EM was used to determine the structure of the hexamer, which is held together by interactions between parasite-specific subunits in the lumenal region. Overall, we identified 17 apicomplexan-specific subunits, and a minimal and nuclear-encoded subunit-a. The hexamer consists of three dimers with an extensive dimer interface that includes bound cardiolipins and the inhibitor IF 1 . Cryo-ET and subtomogram averaging revealed that hexamers arrange into ~20-megadalton pentagonal pyramids in the curved apical membrane regions. Knockout of the linker protein ATPTG11 resulted in the loss of pentagonal pyramids with concomitant aberrantly shaped cristae. Together, this demonstrates that the unique macromolecular arrangement is critical for the maintenance of cristae morphology in Apicomplexa.
Organizational Affiliation:
Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, 17165, Solna, Sweden.