Continuous Assembly of beta-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
Motlova, L., Klimova, N., Fiser, R., Sebo, P., Bumba, L.(2020) J Mol Biol 432: 5696-5710
- PubMed: 32860773
- DOI: https://doi.org/10.1016/j.jmb.2020.08.020
- Primary Citation of Related Structures:
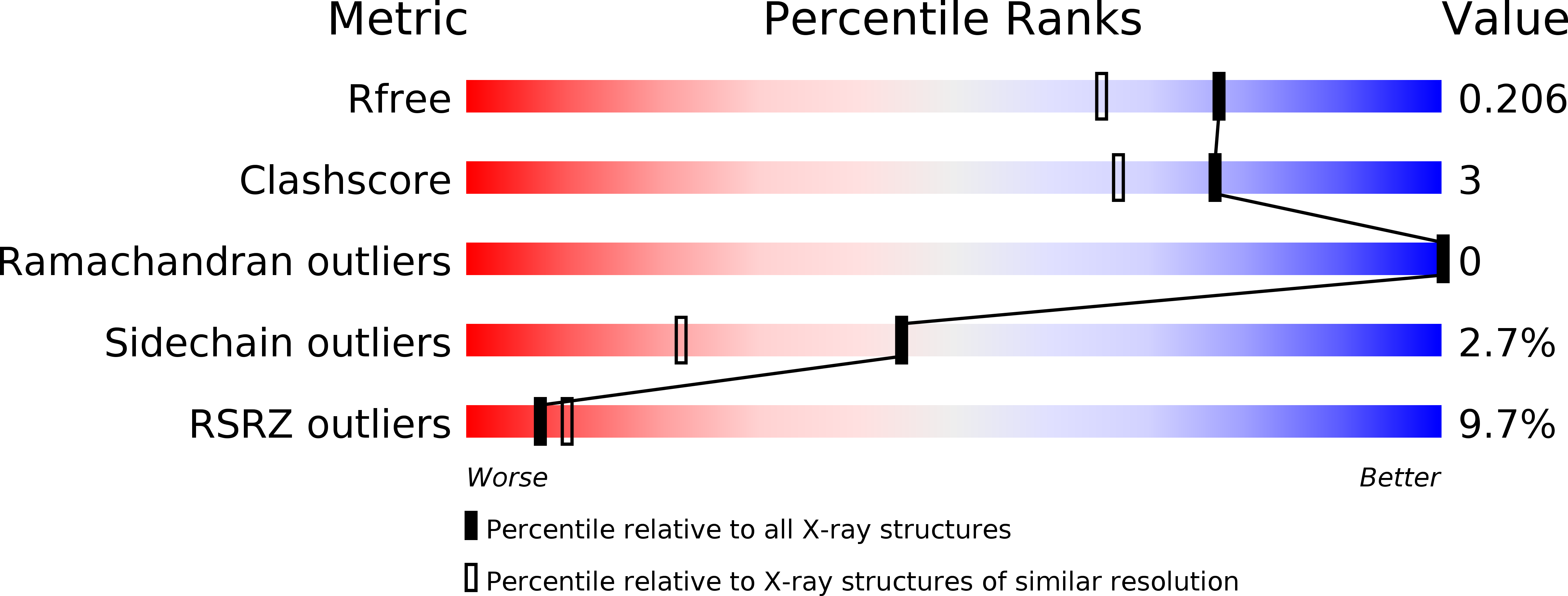
6SUS - PubMed Abstract:
Repeats-in-Toxin (RTX) proteins of Gram-negative bacteria are excreted through the type I secretion system (T1SS) that recognizes non-cleavable C-terminal secretion signals. These are preceded by arrays of glycine and aspartate-rich nonapeptide repeats grouped by four to eight β strands into blocks that fold into calcium-binding parallel β-roll structures. The β-rolls are interspersed by linkers of variable length and sequence and the organization of multiple RTX repeat blocks within large RTX domains remains unknown. Here we examined the structure and function of the RTX domain of Bordetella pertussis adenylate cyclase toxin (CyaA) that is composed of five β-roll RTX blocks. We show that the non-folded RTX repeats maintain the stability of the CyaA polypeptide in the Ca 2+ -depleted bacterial cytosol and thereby enable its efficient translocation through the T1SS apparatus. The efficacy of secretion of truncated CyaA constructs was dictated by the number of retained RTX repeat blocks and depended on the presence of extracellular Ca 2+ ions. We further describe the crystal structure of the RTX blocks IV-V of CyaA (CyaA 1372 - 1681 ) that consists of a contiguous assembly of two β-rolls that differs substantially from the arrangement of the RTX blocks observed in RTX lipases or other RTX proteins. These results provide a novel structural insight into the architecture of the RTX domains of large RTX proteins and support the "push-ratchet" mechanism of the T1SS-mediated secretion of very large RTX proteins.
Organizational Affiliation:
Faculty of Sciences, Charles University, Vinicna 5, 128 44 Prague 2, Czech Republic.