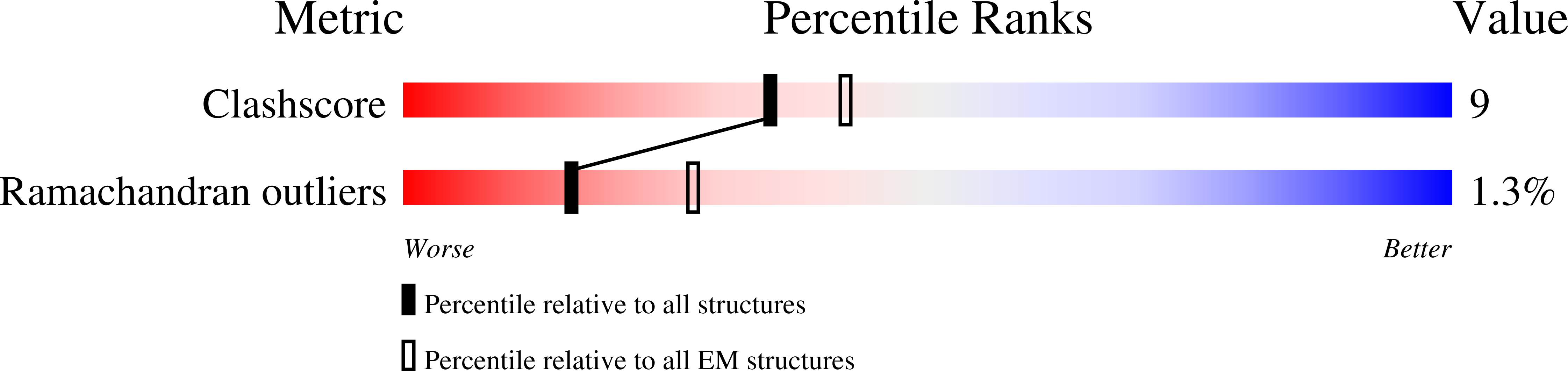Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Horst, B.G., Yokom, A.L., Rosenberg, D.J., Morris, K.L., Hammel, M., Hurley, J.H., Marletta, M.A.(2019) Elife 8
- PubMed: 31566566
- DOI: https://doi.org/10.7554/eLife.50634
- Primary Citation of Related Structures:
6PAS, 6PAT - PubMed Abstract:
Soluble guanylate cyclase (sGC) is the primary receptor for nitric oxide (NO) in mammalian nitric oxide signaling. We determined structures of full-length Manduca sexta sGC in both inactive and active states using cryo-electron microscopy. NO and the sGC-specific stimulator YC-1 induce a 71° rotation of the heme-binding β H-NOX and PAS domains. Repositioning of the β H-NOX domain leads to a straightening of the coiled-coil domains, which, in turn, use the motion to move the catalytic domains into an active conformation. YC-1 binds directly between the β H-NOX domain and the two CC domains. The structural elongation of the particle observed in cryo-EM was corroborated in solution using small angle X-ray scattering (SAXS). These structures delineate the endpoints of the allosteric transition responsible for the major cyclic GMP-dependent physiological effects of NO.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley, Berkeley, United States.