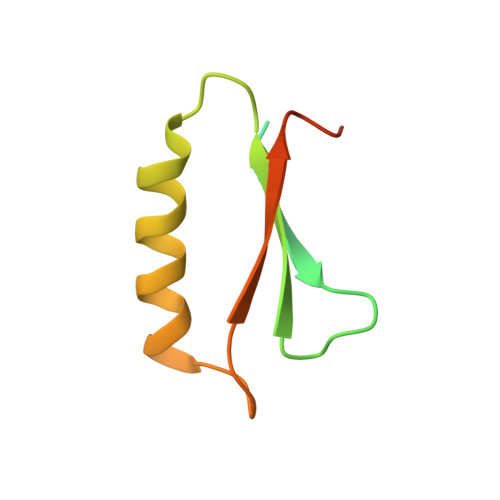
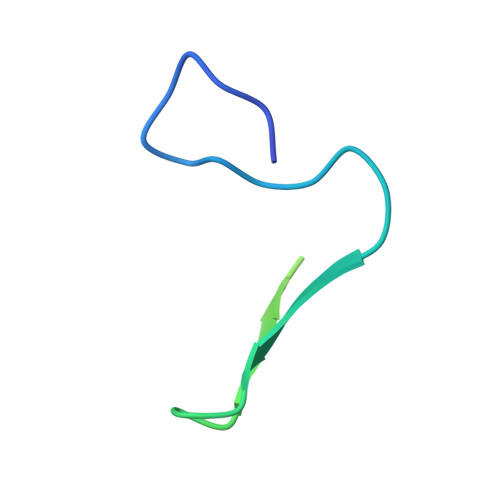
An order-to-disorder structural switch activates the FoxM1 transcription factor.
Marceau, A.H., Brison, C.M., Nerli, S., Arsenault, H.E., McShan, A.C., Chen, E., Lee, H.W., Benanti, J.A., Sgourakis, N.G., Rubin, S.M.(2019) Elife 8
- PubMed: 31134895
- DOI: https://doi.org/10.7554/eLife.46131
- Primary Citation of Related Structures:
6OSW - PubMed Abstract:
Intrinsically disordered transcription factor transactivation domains (TADs) function through structural plasticity, adopting ordered conformations when bound to transcriptional co-regulators. Many transcription factors contain a negative regulatory domain (NRD) that suppresses recruitment of transcriptional machinery through autoregulation of the TAD. We report the solution structure of an autoinhibited NRD-TAD complex within FoxM1, a critical activator of mitotic gene expression. We observe that while both the FoxM1 NRD and TAD are primarily intrinsically disordered domains, they associate and adopt a structured conformation. We identify how Plk1 and Cdk kinases cooperate to phosphorylate FoxM1, which releases the TAD into a disordered conformation that then associates with the TAZ2 or KIX domains of the transcriptional co-activator CBP. Our results support a mechanism of FoxM1 regulation in which the TAD undergoes switching between disordered and different ordered structures.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Cruz, Santa Cruz, United States.