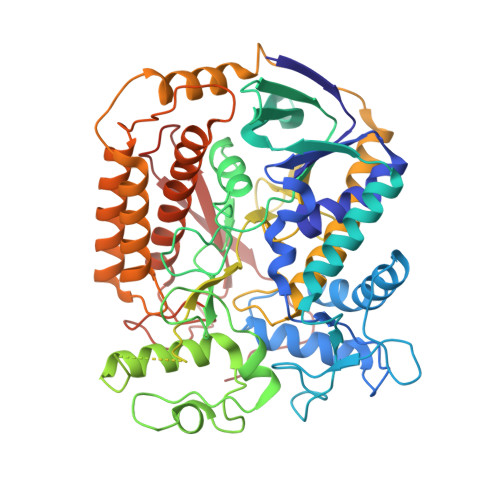
Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Gong, H., Gao, Y., Zhou, X., Xiao, Y., Wang, W., Tang, Y., Zhou, S., Zhang, Y., Ji, W., Yu, L., Tian, C., Lam, S.M., Shui, G., Guddat, L.W., Wong, L.L., Wang, Q., Rao, Z.(2020) Nat Commun 11: 4245-4245
- PubMed: 32843629
- DOI: https://doi.org/10.1038/s41467-020-18011-9
- Primary Citation of Related Structures:
6LUM - PubMed Abstract:
Diheme-containing succinate:menaquinone oxidoreductases (Sdh) are widespread in Gram-positive bacteria but little is known about the catalytic mechanisms they employ for succinate oxidation by menaquinone. Here, we present the 2.8 Å cryo-electron microscopy structure of a Mycobacterium smegmatis Sdh, which forms a trimer. We identified the membrane-anchored SdhF as a subunit of the complex. The 3 kDa SdhF forms a single transmembrane helix and this helix plays a role in blocking the canonically proximal quinone-binding site. We also identified two distal quinone-binding sites with bound quinones. One distal binding site is formed by neighboring subunits of the complex. Our structure further reveals the electron/proton transfer pathway for succinate oxidation by menaquinone. Moreover, this study provides further structural insights into the physiological significance of a trimeric respiratory complex II. The structure of the menaquinone binding site could provide a framework for the development of Sdh-selective anti-mycobacterial drugs.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, Frontiers Science Center for Cell Responses, College of Life Sciences, Nankai University, 300353, Tianjin, China. gonghr@nankai.edu.cn.