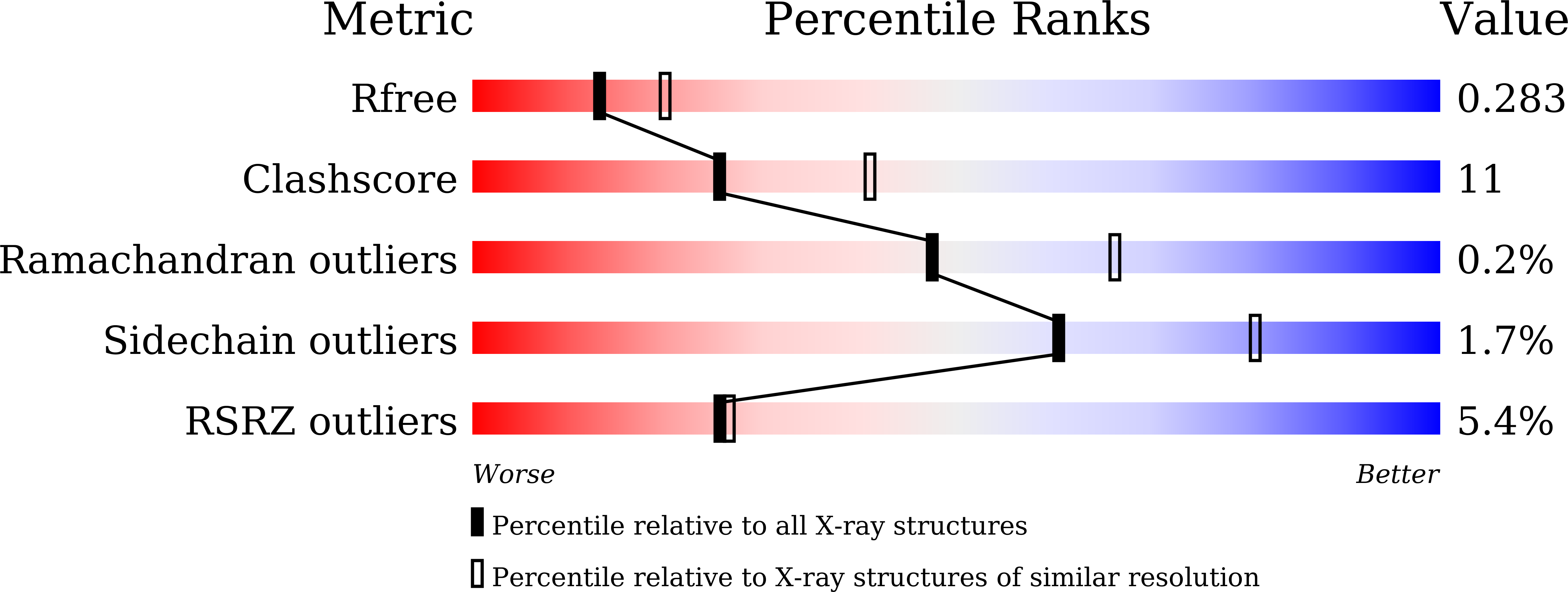
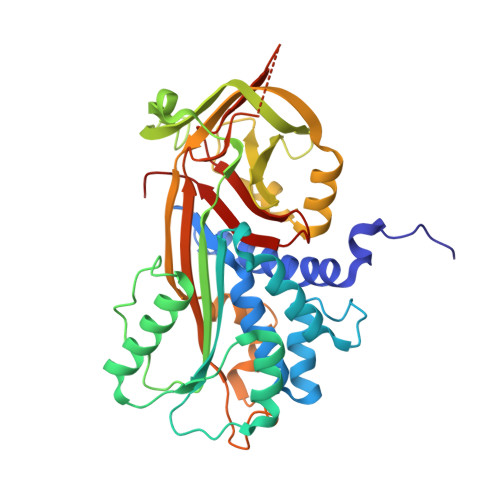
Spatiotemporal regulation of PEDF signaling by type I collagen remodeling.
Kawahara, K., Yoshida, T., Maruno, T., Oki, H., Ohkubo, T., Koide, T., Kobayashi, Y.(2020) Proc Natl Acad Sci U S A 117: 11450-11458
- PubMed: 32385162
- DOI: https://doi.org/10.1073/pnas.2004034117
- Primary Citation of Related Structures:
6LOS - PubMed Abstract:
Dynamic remodeling of the extracellular matrix affects many cellular processes, either directly or indirectly, through the regulation of soluble ligands; however, the mechanistic details of this process remain largely unknown. Here we propose that type I collagen remodeling regulates the receptor-binding activity of pigment epithelium-derived factor (PEDF), a widely expressed secreted glycoprotein that has multiple important biological functions in tissue and organ homeostasis. We determined the crystal structure of PEDF in complex with a disulfide cross-linked heterotrimeric collagen peptide, in which the α(I) chain segments-each containing the respective PEDF-binding region (residues 930 to 938)-are assembled with an α2α1α1 staggered configuration. The complex structure revealed that PEDF specifically interacts with a unique amphiphilic sequence, KGHRGFSGL, of the type I collagen α1 chain, with its proposed receptor-binding sites buried extensively. Molecular docking demonstrated that the PEDF-binding surface of type I collagen contains the cross-link-susceptible Lys930 residue of the α1 chain and provides a good foothold for stable docking with the α1(I) N-telopeptide of an adjacent triple helix in the fibril. Therefore, the binding surface is completely inaccessible if intermolecular crosslinking between two crosslink-susceptible lysyl residues, Lys9 in the N-telopeptide and Lys930, is present. These structural analyses demonstrate that PEDF molecules, once sequestered around newly synthesized pericellular collagen fibrils, are gradually liberated as collagen crosslinking increases, making them accessible for interaction with their target cell surface receptors in a spatiotemporally regulated manner.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Osaka University, Suita, 565-0871 Osaka, Japan.