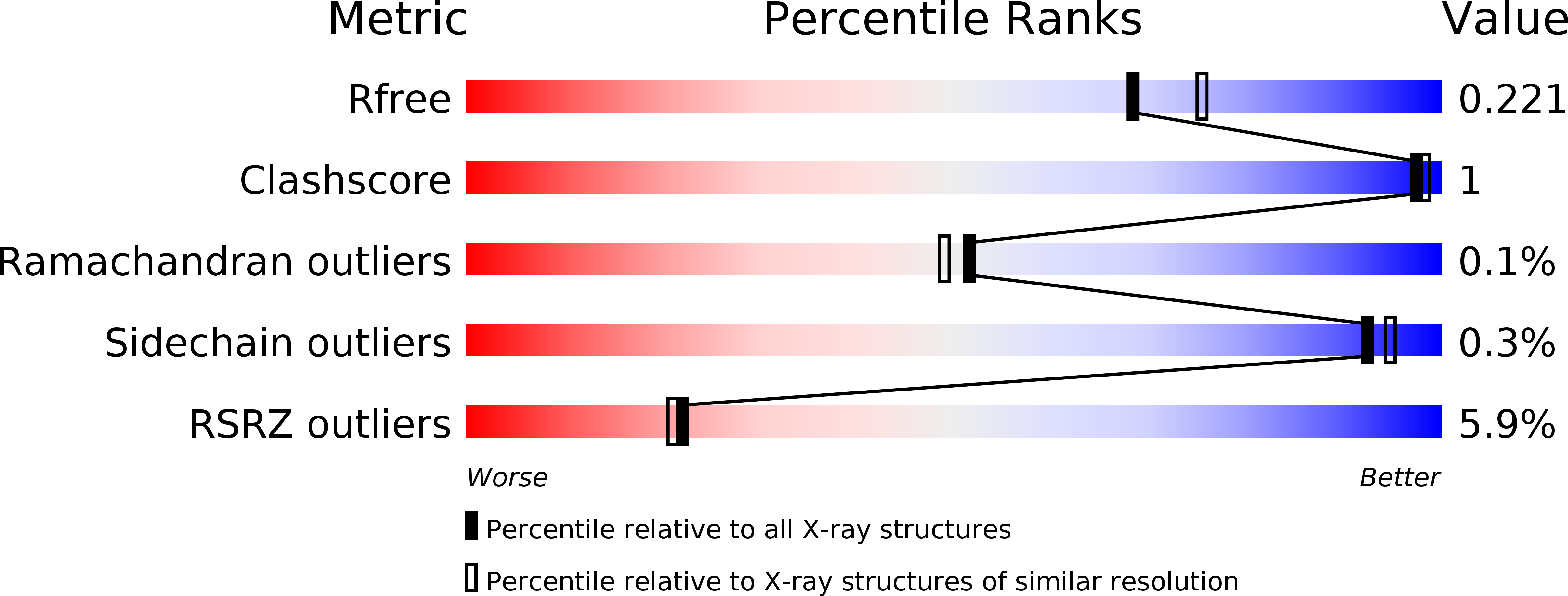
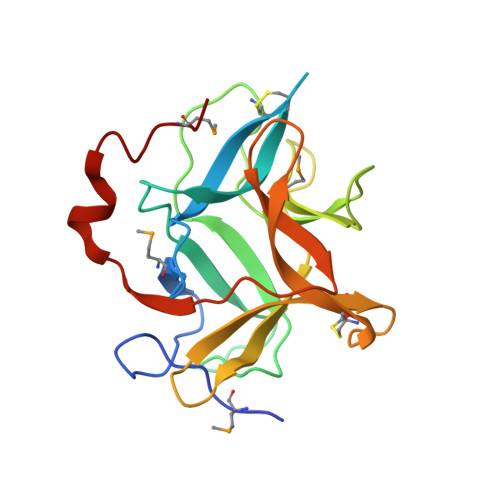
Structural and functional relationship of Cassia obtusifolia trypsin inhibitor to understand its digestive resistance against Pieris rapae.
Zhou, J., Li, C., Chen, A., Zhu, J., Zou, M., Liao, H., Yu, Y.(2020) Int J Biol Macromol 148: 908-920
- PubMed: 31981663
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.01.193
- Primary Citation of Related Structures:
6KV2 - PubMed Abstract:
Although digestive resistance of Kunitz protease inhibitors has been reported extensively, the molecular mechanism is not well established. In the present study, the first X-ray structure of Cassia obtusifolia trypsin inhibitor (COTI), a member of Kunitz protease inhibitors, was solved at a resolution of 1.9 Å. The structure adopted a classic β-trefoil fold with the inhibitory loop protruding from the hydrophobic core. The role of Phe139, a unique residue in Kunitz protease inhibitors, and Arg63 in the COTI structure was verified by F139A and R63E mutants. COTI was a specific inhibitor of bovine trypsin and the result was also verified by COTI-trypsin complex formation. Meanwhile, COTI showed equivalent inhibitory activity with that of soybean trypsin inhibitor against bovine trypsin and midgut trypsin from Pieris rapae. The F139 and R63E mutants further indicated that inhibitory specificity and efficiency of COTI were closely related to the global framework, the conformation and the amino acid composition of reactive loop. Finally, a midgut trypsin from P. rapae (PrSP40), which might be involve in the food digestion, was proposed to be a potential target of COTI and might be a promising target for future crop-protection strategy. The results supported the digestive resistance of COTI.
Organizational Affiliation:
School of Life Science and Engineering, Southwest Jiaotong University, Chengdu, Sichuan, 610031, China. Electronic address: spinezhou@home.swjtu.edu.cn.