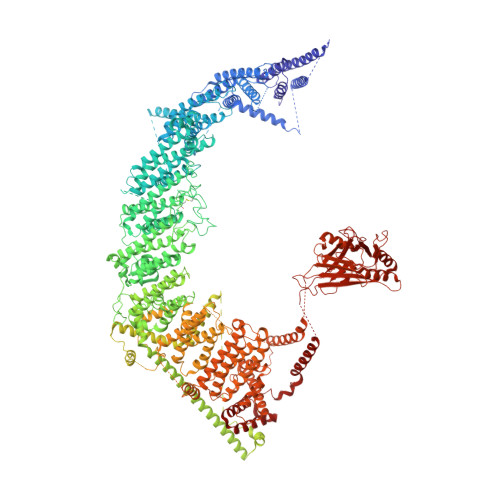
Structure and mechanogating of the mammalian tactile channel PIEZO2.
Wang, L., Zhou, H., Zhang, M., Liu, W., Deng, T., Zhao, Q., Li, Y., Lei, J., Li, X., Xiao, B.(2019) Nature 573: 225-229
- PubMed: 31435011
- DOI: https://doi.org/10.1038/s41586-019-1505-8
- Primary Citation of Related Structures:
6KG7 - PubMed Abstract:
PIEZO2 is a mechanosensitive cation channel that has a key role in sensing touch, tactile pain, breathing and blood pressure. Here we describe the cryo-electron microscopy structure of mouse PIEZO2, which is a three-bladed, propeller-like trimer that comprises 114 transmembrane helices (38 per protomer). Transmembrane helices 1-36 (TM1-36) are folded into nine tandem units of four transmembrane helices each to form the unusual non-planar blades. The three blades are collectively curved into a nano-dome of 28-nm diameter and 10-nm depth, with an extracellular cap-like structure embedded in the centre and a 9-nm-long intracellular beam connecting to the central pore. TM38 and the C-terminal domain are surrounded by the anchor domain and TM37, and enclose the central pore with both transmembrane and cytoplasmic constriction sites. Structural comparison between PIEZO2 and its homologue PIEZO1 reveals that the transmembrane constriction site might act as a transmembrane gate that is controlled by the cap domain. Together, our studies provide insights into the structure and mechanogating mechanism of Piezo channels.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, IDG/McGovern Institute for Brain Research, School of Pharmaceutical Sciences, Tsinghua University, Beijing, China.