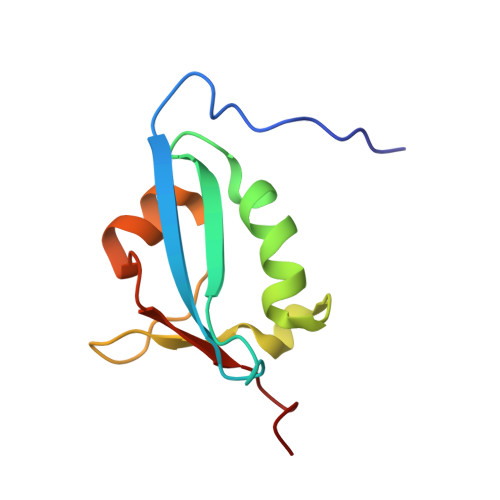
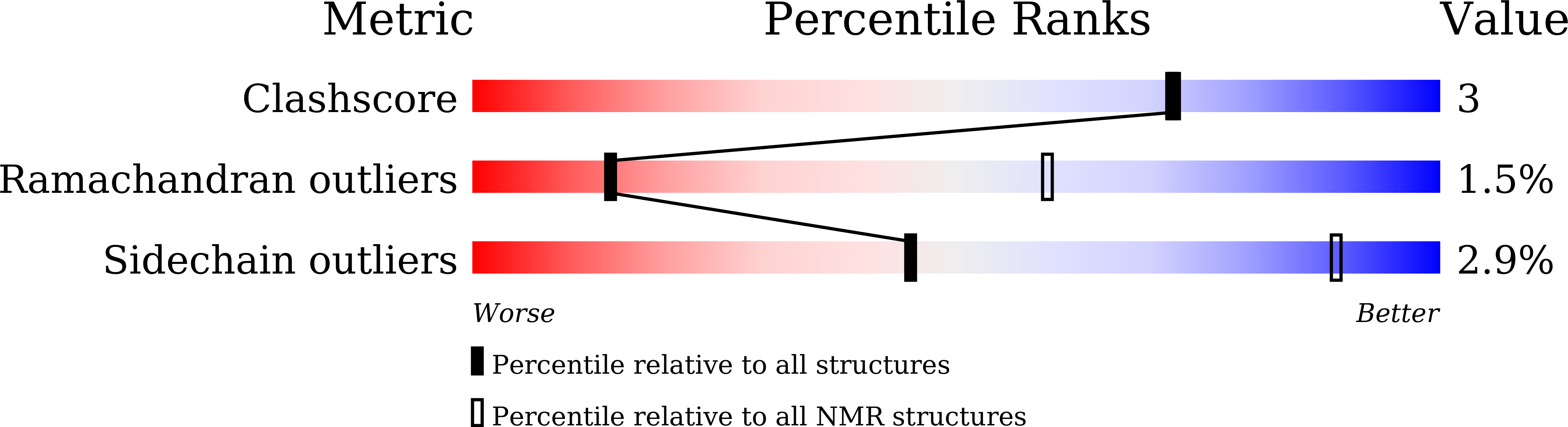Solution structure of TbUfm1 from Trypanosoma brucei and its binding to TbUba5.
Diwu, Y., Zhang, J., Li, M., Yang, X., Shan, F., Ma, H., Zhang, X., Liao, S., Tu, X.(2020) J Struct Biol 212: 107580-107580
- PubMed: 32693018
- DOI: https://doi.org/10.1016/j.jsb.2020.107580
- Primary Citation of Related Structures:
6IZG - PubMed Abstract:
Ubiquitin-like proteins are conserved in eukaryotes and involved in numerous cellular processes. Ufm1 is proved to play important roles in endoplasmic reticulum homeostasis, vesicle transportation and embryonic development. Enzyme cascade of Ufm1 is similar to that of ubiquitin. Mature Ufm1 is activated and conjugated to substrates by assistance of Ufm1 activating enzyme Uba5 (E1), Ufm1 conjugating enzyme Ufc1 (E2), and Ufm1 ligating enzyme Ufl1 (E3). Here, we determined the solution structure of TbUfm1 from Trypanosoma brucei (T. brucei) by NMR spectroscopy and explored the interactions between TbUfm1 and TbUba5/TbUfc1/TbUfl1. TbUfm1 adopts a typical β-grasp fold, which partially wraps a central α-helix and the other two helixes. NMR chemical shift perturbation indicated that TbUfm1 interacts with TbUba5 via a hydrophobic pocket formed by α1α2β1β2. Although the structure and Uba5-interaction mode of TbUfm1 are conserved in Ufm1 proteins, there are also some differences, which might reflect the potential diversity of Ufm1 in evolution and biological functions.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China.