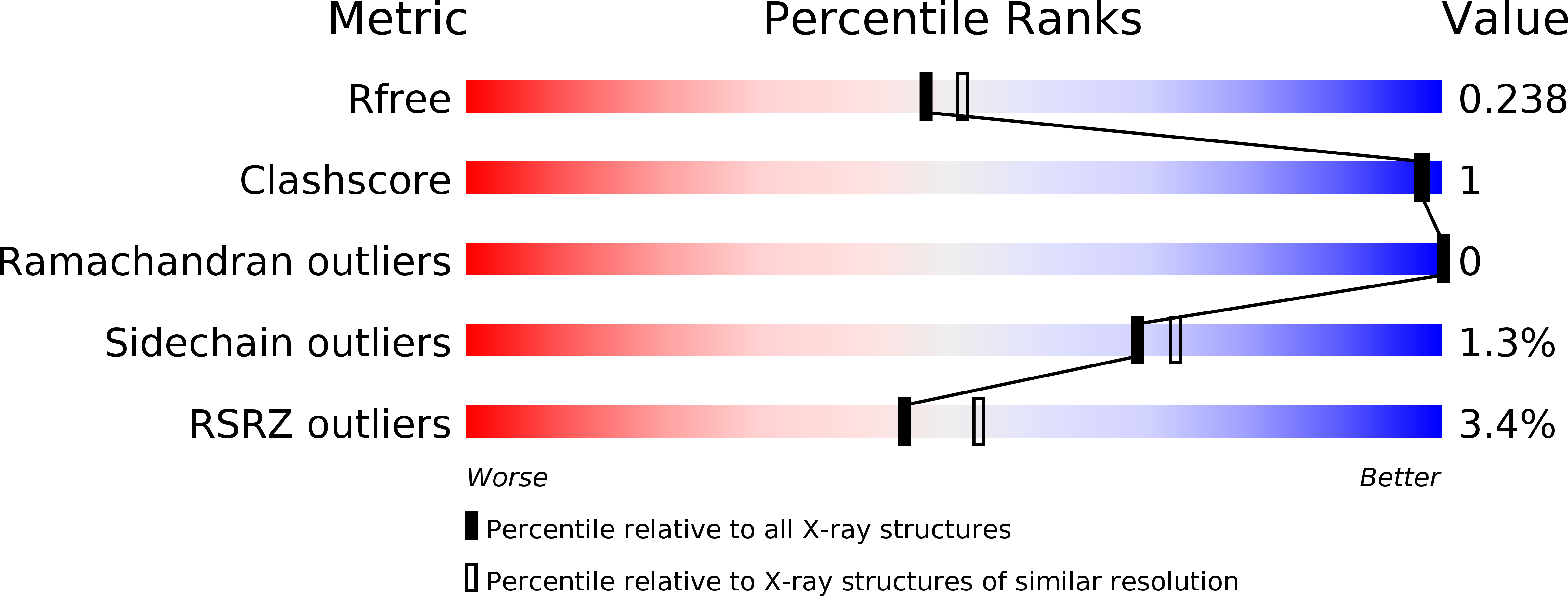
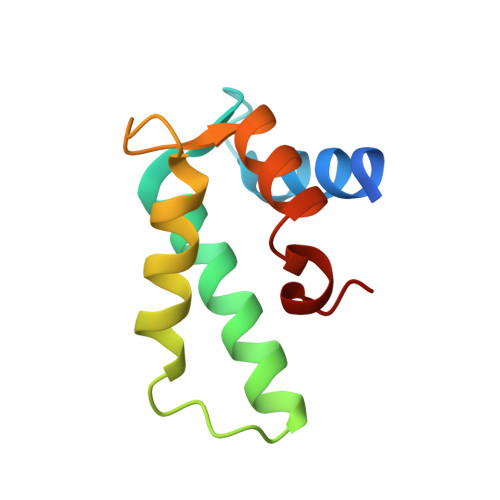
Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Takahashi, D., Suzuki, K., Sakamoto, T., Iwamoto, T., Murata, T., Sakane, F.(2019) Protein Sci 28: 694-706
- PubMed: 30653270
- DOI: https://doi.org/10.1002/pro.3572
- Primary Citation of Related Structures:
6IIE - PubMed Abstract:
Diacylglycerol kinases (DGKs) are multi-domain lipid kinases that phosphorylate diacylglycerol into phosphatidic acid, modulating the levels of these key signaling lipids. Recently, increasing attention has been paid to DGKα isozyme as a potential target for cancer immunotherapy. We have previously shown that DGKα is positively regulated by Ca 2+ binding to its N-terminal EF-hand domains (DGKα-EF). However, little progress has been made for the structural biology of mammalian DGKs and the molecular mechanism underlying the Ca 2+ -triggered activation remains unclear. Here we report the first crystal structure of Ca 2+ -bound DGKα-EF and analyze the structural changes upon binding to Ca 2+ . DGKα-EF adopts a canonical EF-hand fold, but unexpectedly, has an additional α-helix (often called a ligand mimic [LM] helix), which is packed into the hydrophobic core. Biophysical and biochemical analyses reveal that DGKα-EF adopts a protease-susceptible "open" conformation without Ca 2+ that tends to form a dimer. Cooperative binding of two Ca 2+ ions dissociates the dimer into a well-folded monomer, which resists to proteolysis. Taken together, our results provide experimental evidence that Ca 2+ binding induces substantial conformational changes in DGKα-EF, which likely regulates intra-molecular interactions responsible for the activation of DGKα and suggest a possible role of the LM helix for the Ca 2+ -induced conformational changes. SIGNIFICANCE STATEMENT: Diacylglycerol kinases (DGKs), which modulates the levels of two lipid second messengers, diacylglycerol and phosphatidic acid, is still structurally enigmatic enzymes since its first identification in 1959. We here present the first crystal structure of EF-hand domains of diacylglycerol kinase α in its Ca 2+ bound form and characterize Ca 2+ -induced conformational changes, which likely regulates intra-molecular interactions. Our study paves the way for future studies to understand the structural basis of DGK isozymes.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Chiba University, Chiba, Japan.