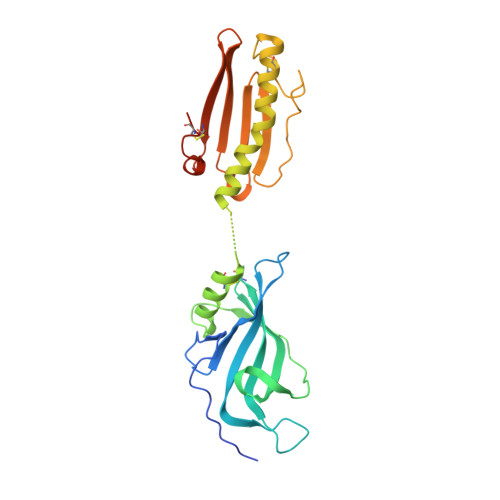
Design and Characterization of Protein E-PilA, a Candidate Fusion Antigen for Nontypeable Haemophilus influenzae Vaccine.
Blais, N., Somers, D., Faubert, D., Labbe, S., Castado, C., Ysebaert, C., Gagnon, L.P., Champagne, J., Gagne, M., Martin, D.(2019) Infect Immun 87
- PubMed: 31085711
- DOI: https://doi.org/10.1128/IAI.00022-19
- Primary Citation of Related Structures:
6GUS, 6GUT - PubMed Abstract:
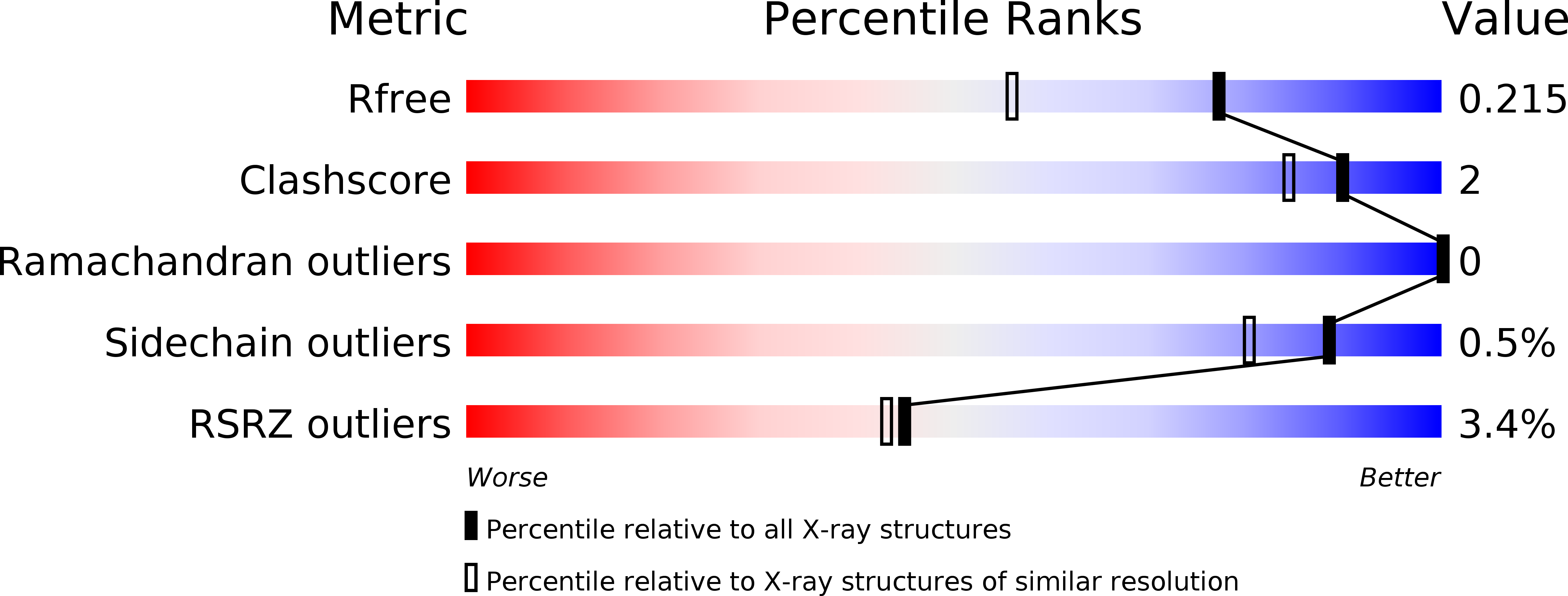
Nontypeable Haemophilus influenzae (NTHi) is a pathogen known for being a frequent cause of acute otitis media in children and respiratory tract infections in adults with chronic obstructive pulmonary disease. In the present study, a vaccine antigen based on the fusion of two known NTHi adhesive proteins, protein E (PE) and a pilin subunit (PilA), was developed. The quality of the combined antigen was investigated through functional, biophysical, and structural analyses. It was shown that the PE and PilA individual structures are not modified in the PE-PilA fusion and that PE-PilA assembles as a dimer in solution, reflecting PE dimerization. PE-PilA was found to bind vitronectin by enzyme-linked immunosorbent assay, as isolated PE does. Disulfide bridges were conserved and homogeneous, which was determined by peptide mapping and top-down analysis of PE, PilA, and PE-PilA molecules. Finally, the PE-PilA crystal showed a PE entity with a three-dimensional (3D) structure similar to that of the recently published isolated PE, while the structure of the PilA entity was similar to that of a 3D model elaborated from two other type 4 pilin subunits. Taken together, our observations suggest that the two tethered proteins behave independently within the chimeric molecule and display structures similar to those of the respective isolated antigens, which are important characteristics for eliciting optimal antibody-mediated immunity. PE and PilA can thus be further developed as a single fusion protein in a vaccine perspective, in the knowledge that tethering the two antigens does not perceptibly compromise the structural attributes offered by the individual antigens.
Organizational Affiliation:
GSK, Rixensart, Belgium Normand.x.blais@gsk.com.