African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Pinger, J., Nesic, D., Ali, L., Aresta-Branco, F., Lilic, M., Chowdhury, S., Kim, H.S., Verdi, J., Raper, J., Ferguson, M.A.J., Papavasiliou, F.N., Stebbins, C.E.(2018) Nat Microbiol 3: 932-938
- PubMed: 29988048
- DOI: https://doi.org/10.1038/s41564-018-0187-6
- Primary Citation of Related Structures:
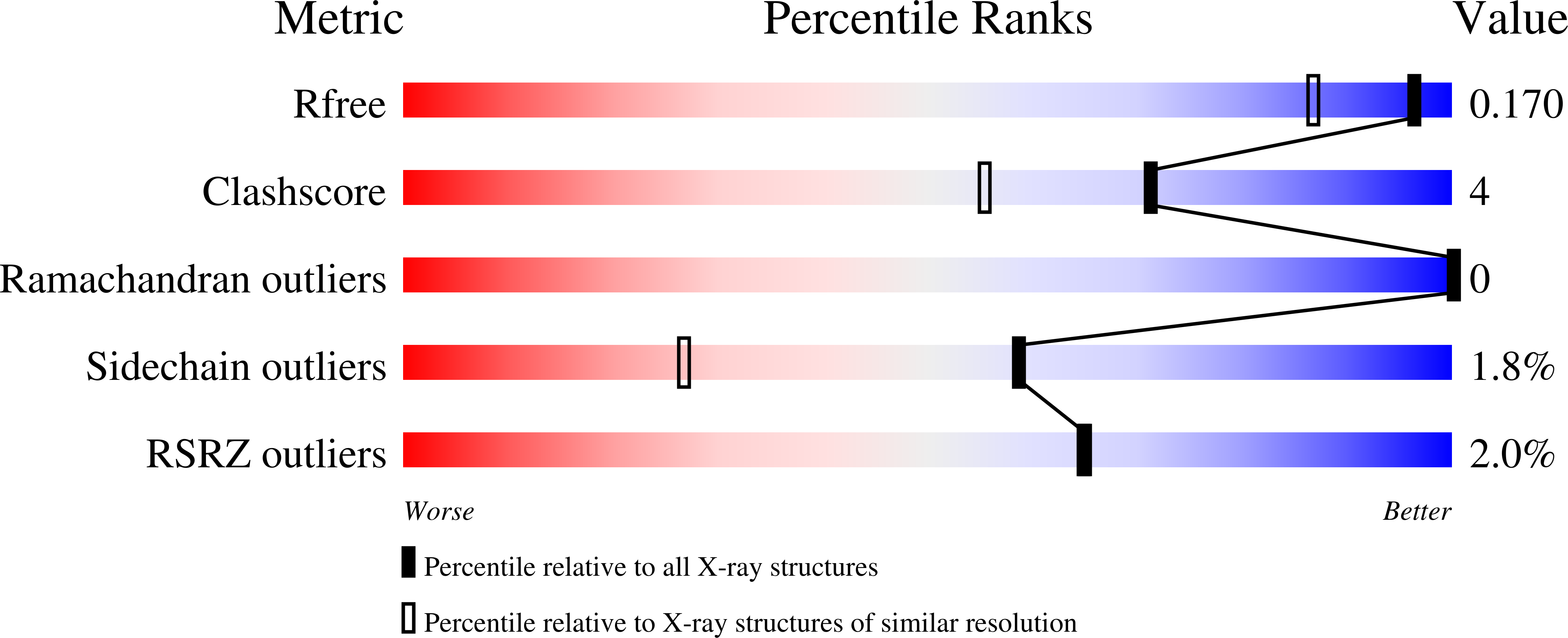
6ELC - PubMed Abstract:
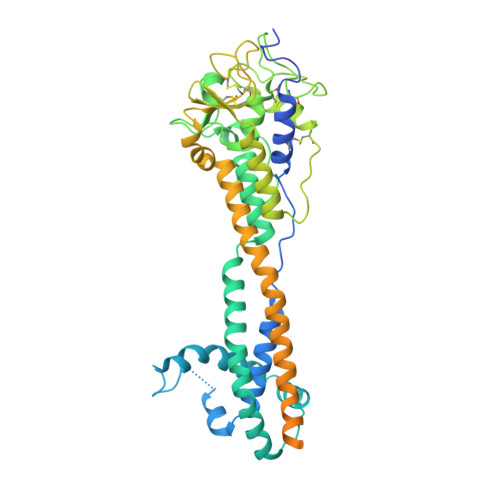
The African trypanosome Trypanosoma brucei spp. is a paradigm for antigenic variation, the orchestrated alteration of cell surface molecules to evade host immunity. The parasite elicits robust antibody-mediated immune responses to its variant surface glycoprotein (VSG) coat, but evades immune clearance by repeatedly accessing a large genetic VSG repertoire and 'switching' to antigenically distinct VSGs. This persistent immune evasion has been ascribed exclusively to amino-acid variance on the VSG surface presented by a conserved underlying protein architecture. We establish here that this model does not account for the scope of VSG structural and biochemical diversity. The 1.4-Å-resolution crystal structure of the variant VSG3 manifests divergence in the tertiary fold and oligomeric state. The structure also reveals an O-linked carbohydrate on the top surface of VSG3. Mass spectrometric analysis indicates that this O-glycosylation site is heterogeneously occupied in VSG3 by zero to three hexose residues and is also present in other VSGs. We demonstrate that this O-glycosylation increases parasite virulence by impairing the generation of protective immunity. These data alter the paradigm of antigenic variation by the African trypanosome, expanding VSG variability beyond amino-acid sequence to include surface post-translational modifications with immunomodulatory impact.
Organizational Affiliation:
The Rockefeller University, Laboratory of Lymphocyte Biology, New York, NY, USA.