PRISEs (progesterone 5 beta-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism.
Schmidt, K., Petersen, J., Munkert, J., Egerer-Sieber, C., Hornig, M., Muller, Y.A., Kreis, W.(2018) Phytochemistry 156: 9-19
- PubMed: 30172078
- DOI: https://doi.org/10.1016/j.phytochem.2018.08.012
- Primary Citation of Related Structures:
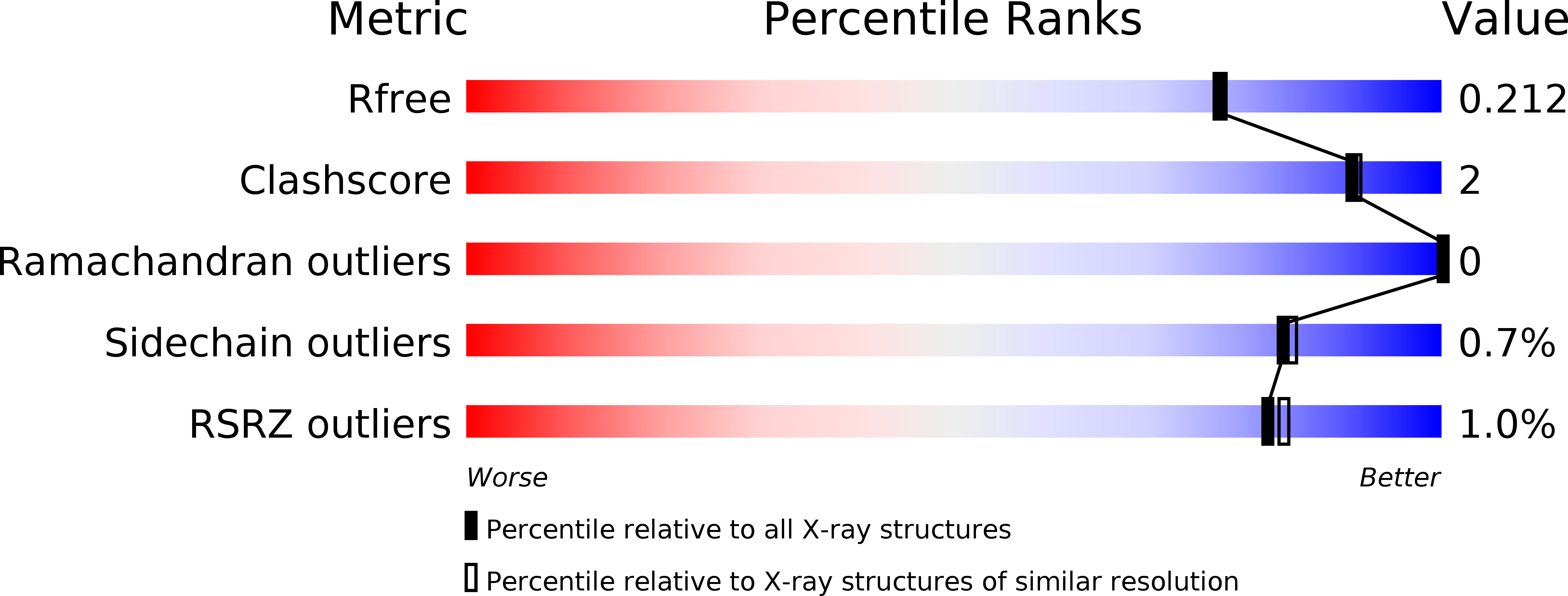
6EL3 - PubMed Abstract:
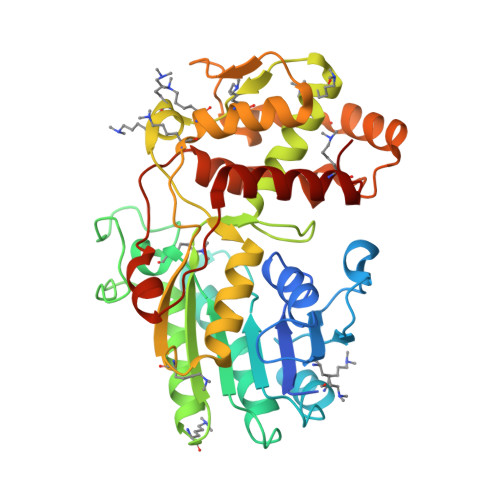
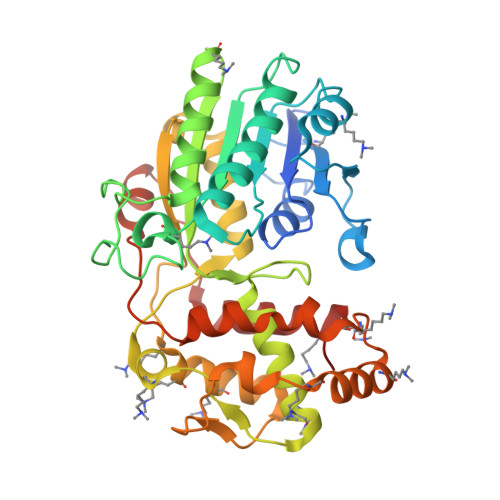
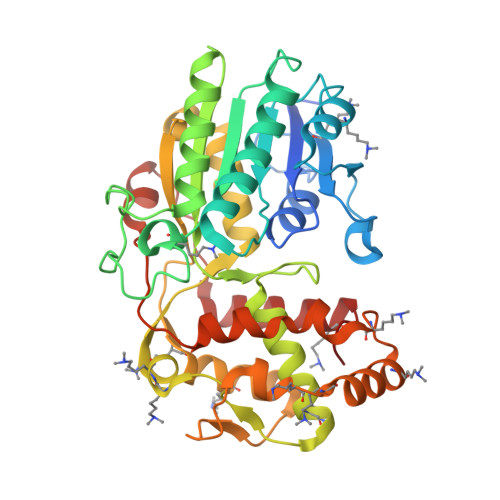
PRISEs (progesterone 5β-reductase and/or iridoid synthase-like 1,4-enone reductases) are involved in cardenolide and iridoid biosynthesis. We here investigated a PRISE (rAtSt5βR) from Arabidopsis thaliana, a plant producing neither cardenolides nor iridoids. The structure of rAtSt5βR was elucidated with X-ray crystallography and compared to the known structures of PRISEs from Catharanthus roseus (rCrISY) and Digitalis lanata (rDlP5βR). The three enzymes show a high degree of sequence and structure conservation in the active site. Amino acids previously considered to allow discrimination between progesterone 5β-reductase and iridoid synthase were interchanged among rAtSt5βR, rCrISY and rDlP5βR applying site-directed mutagenesis. Structural homologous substitutions had different effects, and changes in progesterone 5β-reductase and iridoid synthase activity were not correlated in all cases. Our results help to explain fortuitous emergence of metabolic pathways and product accumulation. The fact that PRISEs are found ubiquitously in spermatophytes insinuates that PRISEs might have a more general function in plant metabolism such as, for example, the detoxification of reactive carbonyl species.
Organizational Affiliation:
Division of Biotechnology, Department of Biology, Friedrich-Alexander-University Erlangen-Nürnberg, D-91054 Erlangen, Germany.