A Viral Protein Restricts Drosophila RNAi Immunity by Regulating Argonaute Activity and Stability.
Nayak, A., Kim, D.Y., Trnka, M.J., Kerr, C.H., Lidsky, P.V., Stanley, D.J., Rivera, B.M., Li, K.H., Burlingame, A.L., Jan, E., Frydman, J., Gross, J.D., Andino, R.(2018) Cell Host Microbe 24: 542-557.e9
- PubMed: 30308158
- DOI: https://doi.org/10.1016/j.chom.2018.09.006
- Primary Citation of Related Structures:
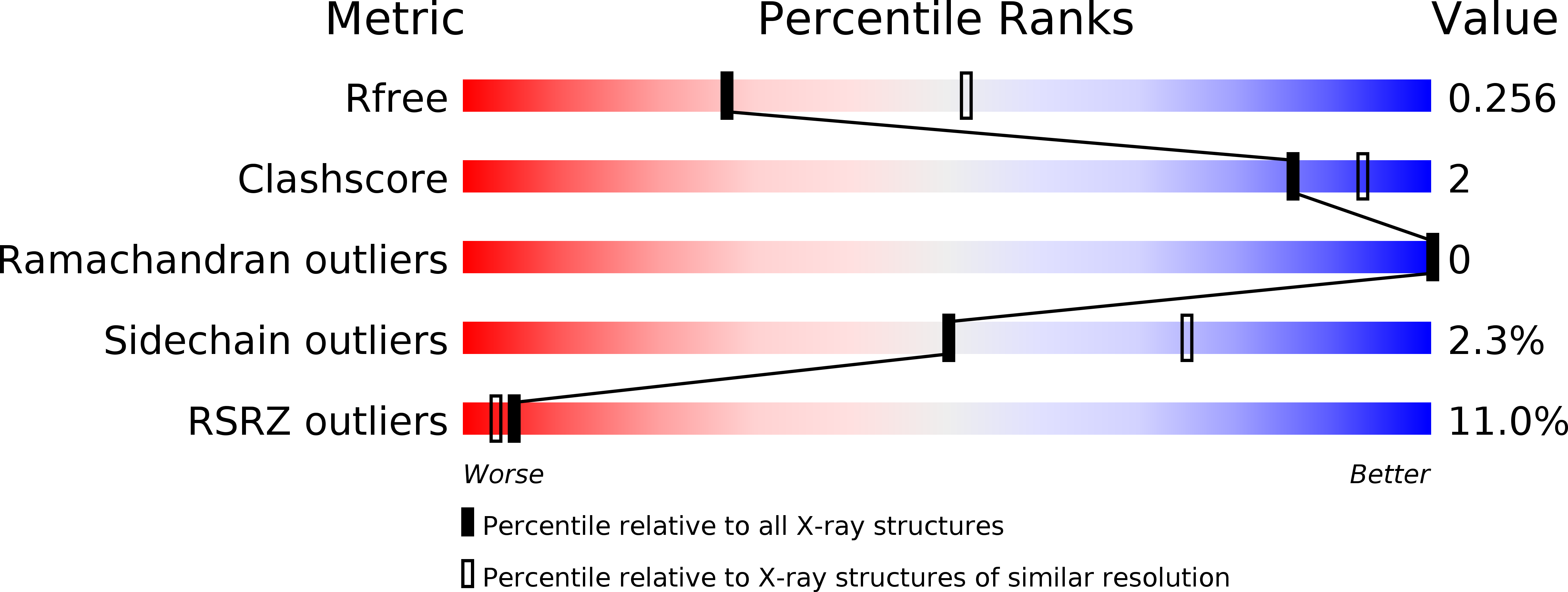
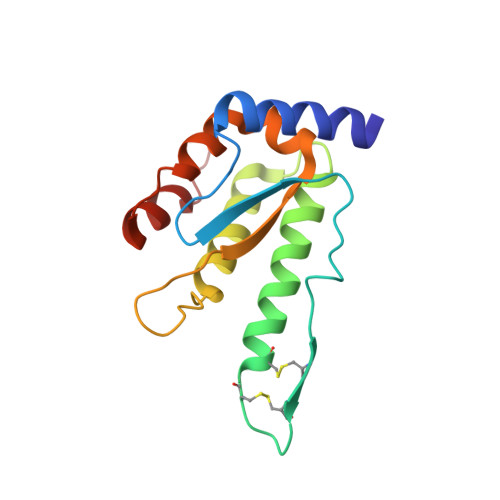
6C3R - PubMed Abstract:
The dicistrovirus, Cricket paralysis virus (CrPV) encodes an RNA interference (RNAi) suppressor, 1A, which modulates viral virulence. Using the Drosophila model, we combined structural, biochemical, and virological approaches to elucidate the strategies by which CrPV-1A restricts RNAi immunity. The atomic resolution structure of CrPV-1A uncovered a flexible loop that interacts with Argonaute 2 (Ago-2), thereby inhibiting Ago-2 endonuclease-dependent immunity. Mutations disrupting Ago-2 binding attenuates viral pathogenesis in wild-type but not Ago-2-deficient flies. CrPV-1A also contains a BC-box motif that enables the virus to hijack a host Cul2-Rbx1-EloBC ubiquitin ligase complex, which promotes Ago-2 degradation and virus replication. Our study uncovers a viral-based dual regulatory program that restricts antiviral immunity by direct interaction with and modulation of host proteins. While the direct inhibition of Ago-2 activity provides an efficient mechanism to establish infection, the recruitment of a ubiquitin ligase complex enables CrPV-1A to amplify Ago-2 inactivation to restrict further antiviral RNAi immunity.
Organizational Affiliation:
Department of Microbiology and Immunology, University of California San Francisco, San Francisco, CA 94158, USA; Department of Biology and Genetics, Stanford University, Stanford, CA 94305, USA.