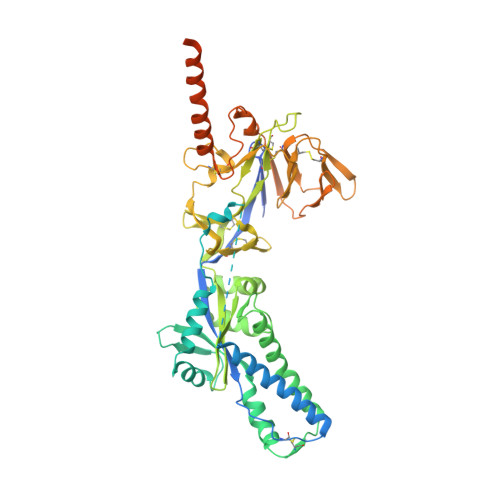
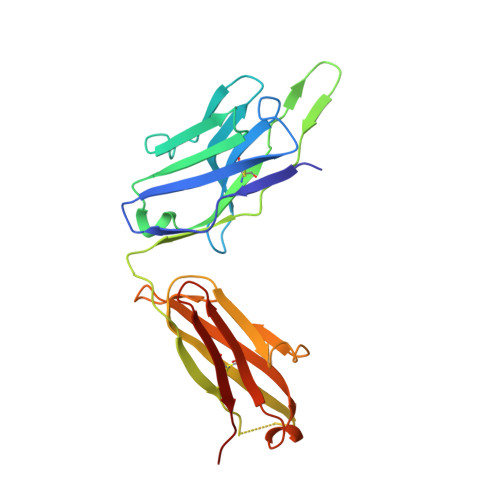
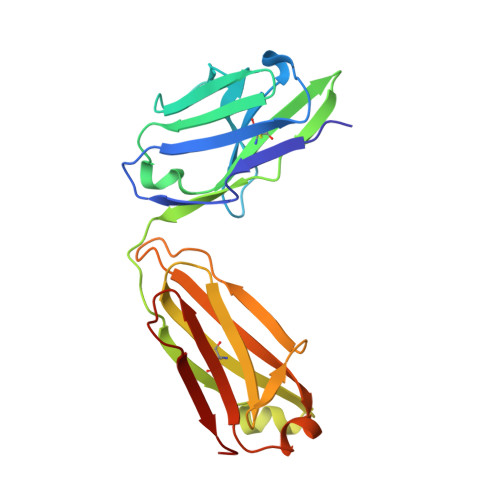
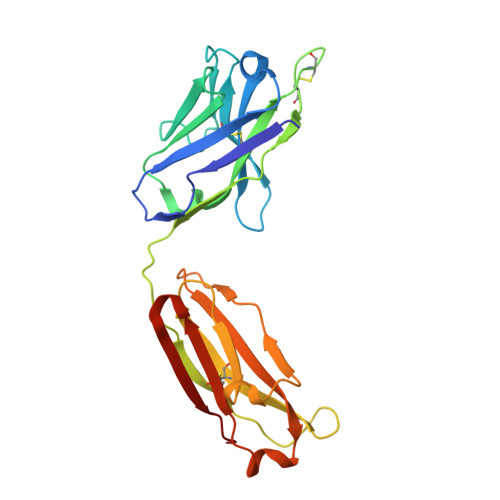
Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Goodwin, E., Gilman, M.S.A., Wrapp, D., Chen, M., Ngwuta, J.O., Moin, S.M., Bai, P., Sivasubramanian, A., Connor, R.I., Wright, P.F., Graham, B.S., McLellan, J.S., Walker, L.M.(2018) Immunity 48: 339-349.e5
- PubMed: 29396163
- DOI: https://doi.org/10.1016/j.immuni.2018.01.005
- Primary Citation of Related Structures:
6APB, 6APC, 6APD - PubMed Abstract:
Respiratory syncytial virus (RSV) is a leading cause of infant mortality, and there are currently no licensed vaccines to protect this vulnerable population. A comprehensive understanding of infant antibody responses to natural RSV infection would facilitate vaccine development. Here, we isolated more than 450 RSV fusion glycoprotein (F)-specific antibodies from 7 RSV-infected infants and found that half of the antibodies recognized only two antigenic sites. Antibodies targeting both sites showed convergent sequence features, and structural studies revealed the molecular basis for their recognition of RSV F. A subset of antibodies targeting one of these sites displayed potent neutralizing activity despite lacking somatic mutations, and similar antibodies were detected in RSV-naive B cell repertoires, suggesting that expansion of these B cells in infants may be possible with suitably designed vaccine antigens. Collectively, our results provide fundamental insights into infant antibody responses and a framework for the rational design of age-specific RSV vaccines.
Organizational Affiliation:
Adimab LLC, Lebanon, NH 03766, USA.