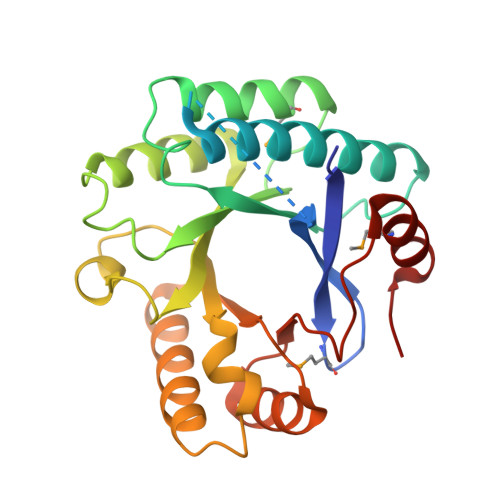
The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP inMycobacterium smegmatis.
Chen, H.J., Li, N., Luo, Y., Jiang, Y.L., Zhou, C.Z., Chen, Y., Li, Q.(2018) Biochem J 475: 1295-1308
- PubMed: 29555845
- DOI: https://doi.org/10.1042/BCJ20180079
- Primary Citation of Related Structures:
5YRP - PubMed Abstract:
The second messenger c-di-GMP [bis-(3'-5')-cyclic dimeric guanosine monophosphate] plays a key role in bacterial growth, survival and pathogenesis, and thus its intracellular homeostasis should be finely maintained. Mycobacterium smegmatis encodes a GAF (mammalian c G MP-regulated phosphodiesterases, Anabaena a denylyl cyclases and Escherichia coli transcription activator F hlA) domain containing bifunctional enzyme DcpA ( d iguanylate c yclase and p hosphodiesterase A ) that catalyzes the synthesis and hydrolysis of c-di-GMP . Here, we found that M. smegmatis DcpA catalyzes the hydrolysis of c-di-GMP at a higher velocity, compared with synthetic activity, resulting in a sum reaction from the ultimate substrate GTP to the final product pGpG [5'-phosphoguanylyl-(3'-5')-guanosine]. Fusion with the N-terminal GAF domain enables the GGDEF (Gly-Gly-Asp-Glu-Phe) domain of DcpA to dimerize and accordingly gain synthetic activity. Screening of putative metabolites revealed that GDP is the ligand of the GAF domain. Binding of GDP to the GAF domain down-regulates synthetic activity, but up-regulates hydrolytic activity, which, in consequence, might enable a timely response to the transient accumulation of c-di-GMP at the stationary phase or under stresses. Combined with the crystal structure of the EAL (Glu-Ala-Leu) domain and the small-angle X-ray scattering data, we propose a putative regulatory model of the GAF domain finely tuned by the intracellular GTP/GDP ratio. These findings help us to better understand the concerted control of the synthesis and hydrolysis of c-di-GMP in M. smegmatis in various microenvironments.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China.