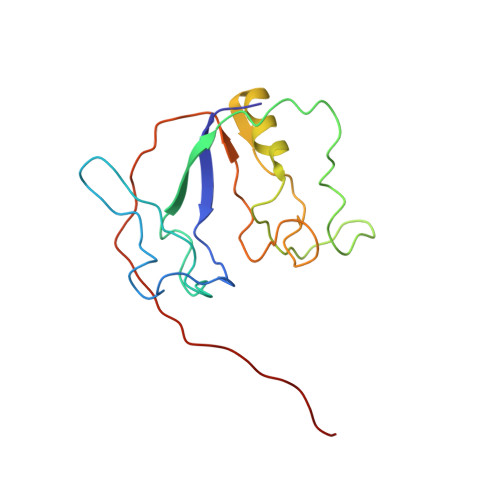
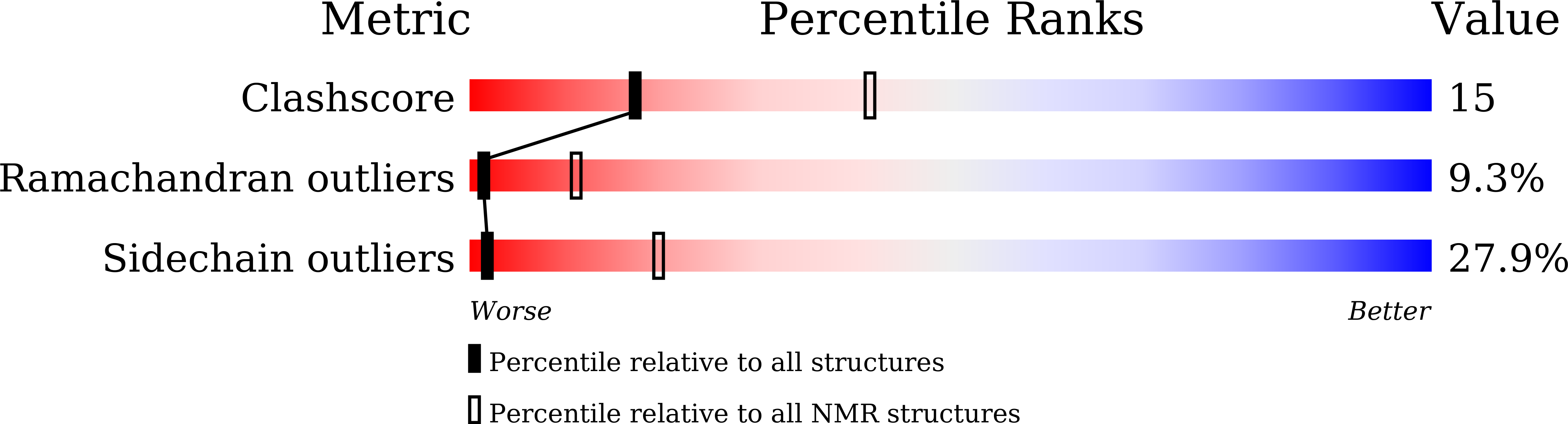Solution NMR Structure and Backbone Dynamics of Partially Disordered Arabidopsis thaliana Phloem Protein 16-1, a Putative mRNA Transporter.
Sashi, P., Singarapu, K.K., Bhuyan, A.K.(2018) Biochemistry 57: 912-924
- PubMed: 29320165
- DOI: https://doi.org/10.1021/acs.biochem.7b01071
- Primary Citation of Related Structures:
5YQ3 - PubMed Abstract:
Although RNA-binding proteins in plant phloem are believed to perform long-distance systemic transport of RNA in the phloem conduit, the structure of none of them is known. Arabidopsis thaliana phloem protein 16-1 (AtPP16-1) is such a putative mRNA transporter whose structure and backbone dynamics have been studied at pH 4.1 and 25 °C by high-resolution nuclear magnetic resonance spectroscopy. Results obtained using basic optical spectroscopic tools show that the protein is unstable with little secondary structure near the physiological pH of the phloem sap. Fluorescence-monitored titrations reveal that AtPP16-1 binds not only A. thaliana RNA (K diss ∼ 67 nM) but also sheared DNA and model dodecamer DNA, though the affinity for DNA is ∼15-fold lower. In the solution structure of the protein, secondary structural elements are formed by residues 3-9 (β1), 56-62 (β2), 133-135 (β3), and 96-110 (α-helix). Most of the rest of the chain segments are disordered. The N-terminally disordered regions (residues 10-55) form a small lobe, which conjoins the rest of the molecule via a deep and large irregular cleft that could have functional implications. The average order parameter extracted by model-free analysis of 15 N relaxation and { 1 H}- 15 N heteronuclear NOE data is 0.66, suggesting less restricted backbone motion. The average conformational entropy of the backbone NH vectors is -0.31 cal mol -1 K -1 . These results also suggest structural disorder in AtPP16-1.
Organizational Affiliation:
School of Chemistry, University of Hyderabad , Hyderabad 500046, India.