Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch
Zhu, K., Shan, Z., Chen, X., Cai, Y., Cui, L., Yao, W., Wang, Z., Shi, P., Tian, C., Lou, J., Xie, Y., Wen, W.(2017) EMBO Rep 18: 1618-1630
- PubMed: 28747490
- DOI: https://doi.org/10.15252/embr.201744454
- Primary Citation of Related Structures:
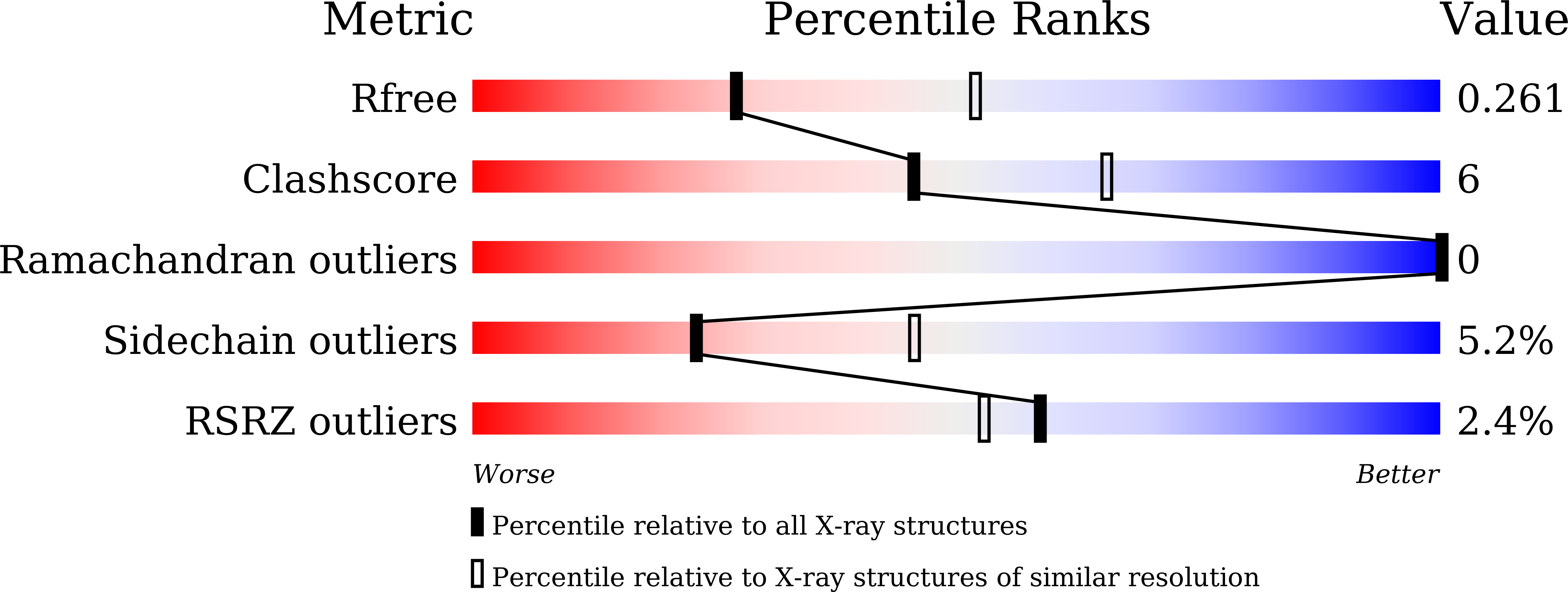
5XMC - PubMed Abstract:
The Nedd4 family E3 ligases are key regulators of cell growth and proliferation and are often misregulated in human cancers and other diseases. The ligase activities of Nedd4 E3s are tightly controlled via auto-inhibition. However, the molecular mechanism underlying Nedd4 E3 auto-inhibition and activation is poorly understood. Here, we show that the WW domains proceeding the catalytic HECT domain play an inhibitory role by binding directly to HECT in the Nedd4 E3 family member Itch. Our structural and biochemical analyses of Itch reveal that the WW2 domain and a following linker allosterically lock HECT in an inactive state inhibiting E2-E3 transthiolation. Binding of the Ndfip1 adaptor or JNK1-mediated phosphorylation relieves the auto-inhibition of Itch in a WW2-dependent manner. Aberrant activation of Itch leads to migration defects of cortical neurons during development. Our study provides a new mechanism governing the regulation of Itch.
Organizational Affiliation:
Department of Neurosurgery, Huashan Hospital, Institutes of Biomedical Sciences, Fudan University, Shanghai, China.