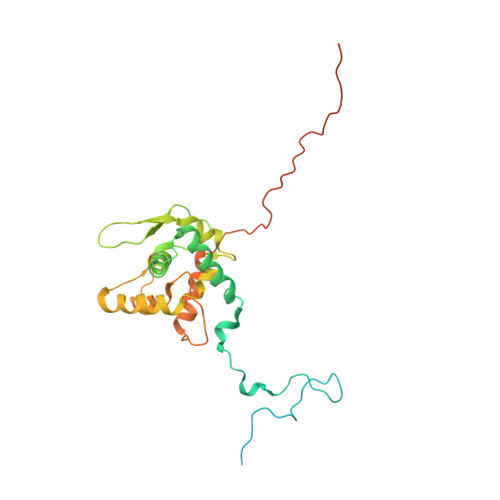
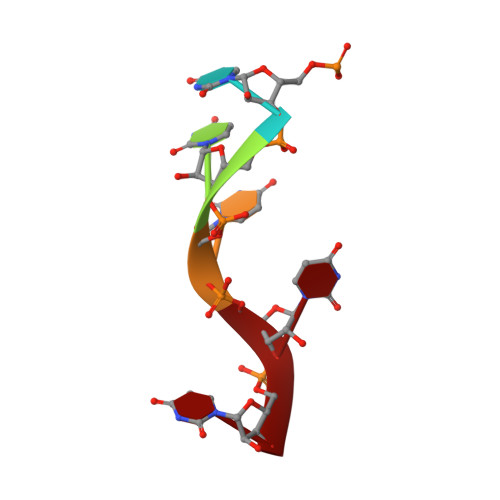
Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses.
Zamora, M., Mendez-Lopez, E., Agirrezabala, X., Cuesta, R., Lavin, J.L., Sanchez-Pina, M.A., Aranda, M.A., Valle, M.(2017) Sci Adv 3: eaao2182-eaao2182
- PubMed: 28948231
- DOI: https://doi.org/10.1126/sciadv.aao2182
- Primary Citation of Related Structures:
5ODV - PubMed Abstract:
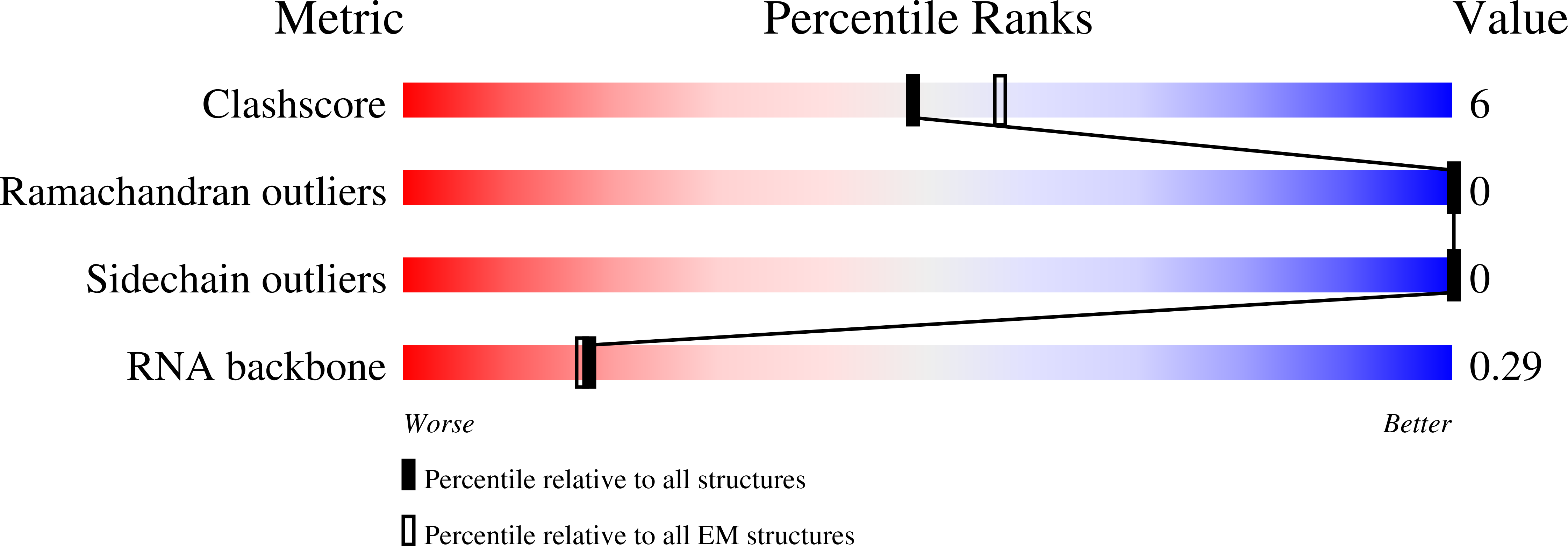
Potyviruses constitute the second largest genus of plant viruses and cause important economic losses in a large variety of crops; however, the atomic structure of their particles remains unknown. Infective potyvirus virions are long flexuous filaments where coat protein (CP) subunits assemble in helical mode bound to a monopartite positive-sense single-stranded RNA [(+)ssRNA] genome. We present the cryo-electron microscopy (cryoEM) structure of the potyvirus watermelon mosaic virus at a resolution of 4.0 Å. The atomic model shows a conserved fold for the CPs of flexible filamentous plant viruses, including a universally conserved RNA binding pocket, which is a potential target for antiviral compounds. This conserved fold of the CP is widely distributed in eukaryotic viruses and is also shared by nucleoproteins of enveloped viruses with segmented (-)ssRNA (negative-sense ssRNA) genomes, including influenza viruses.
Organizational Affiliation:
Molecular Recognition and Host-Pathogen Interactions, Center for Cooperative Research in Biosciences, CIC bioGUNE, 48160 Derio, Spain.