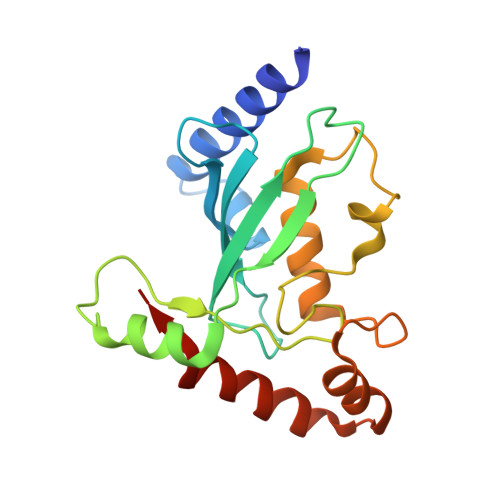
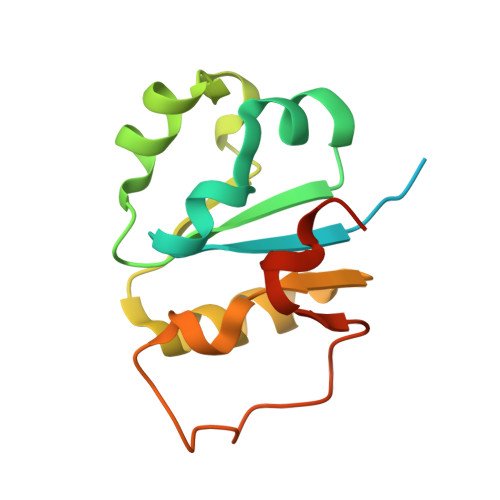
Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Groves, M.R., Schroer, C.F.E., Middleton, A.J., Lunev, S., Danda, N., Ali, A.M., Marrink, S.J., Williams, C.(2018) Biochem Biophys Res Commun 496: 562-567
- PubMed: 29288668
- DOI: https://doi.org/10.1016/j.bbrc.2017.12.150
- Primary Citation of Related Structures:
5NKZ, 5NL8 - PubMed Abstract:
Pex4p is a peroxisomal E2 involved in ubiquitinating the conserved cysteine residue of the cycling receptor protein Pex5p. Previously, we demonstrated that Pex4p from the yeast Saccharomyces cerevisiae binds directly to the peroxisomal membrane protein Pex22p and that this interaction is vital for receptor ubiquitination. In addition, Pex22p binding allows Pex4p to specifically produce lysine 48 linked ubiquitin chains in vitro through an unknown mechanism. This activity is likely to play a role in targeting peroxisomal proteins for proteasomal degradation. Here we present the crystal structures of Pex4p alone and in complex with Pex22p from the yeast Hansenula polymorpha. Comparison of the two structures demonstrates significant differences to the active site of Pex4p upon Pex22p binding while molecular dynamics simulations suggest that Pex22p binding facilitates active site remodelling of Pex4p through an allosteric mechanism. Taken together, our data provide insights into how Pex22p binding allows Pex4p to build K48-linked Ub chains.
Organizational Affiliation:
Department of Drug Design, Groningen Research Institute of Pharmacy, University of Groningen, 9713AV, The Netherlands.