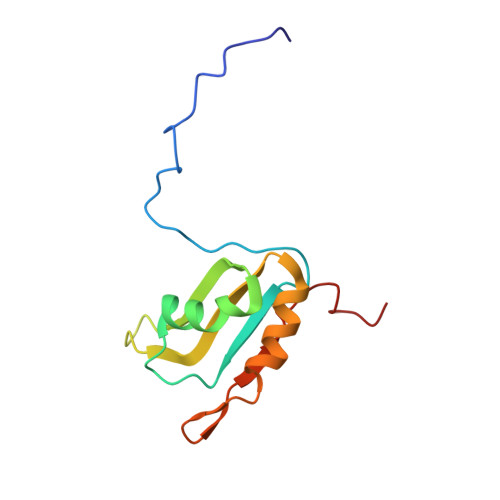
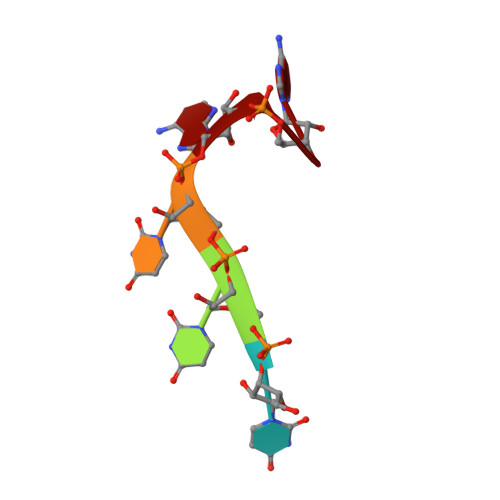
Aromatic side-chain conformational switch on the surface of the RNA Recognition Motif enables RNA discrimination.
Diarra Dit Konte, N., Krepl, M., Damberger, F.F., Ripin, N., Duss, O., Sponer, J., Allain, F.H.(2017) Nat Commun 8: 654-654
- PubMed: 28935965
- DOI: https://doi.org/10.1038/s41467-017-00631-3
- Primary Citation of Related Structures:
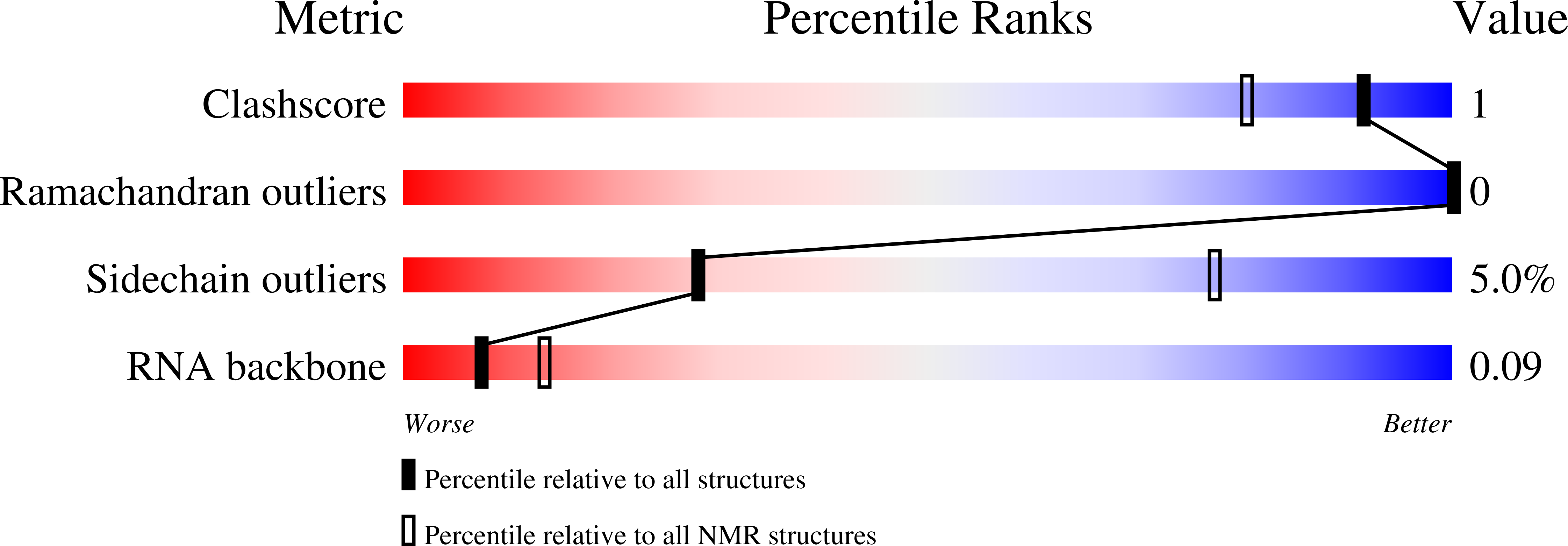
5M8I - PubMed Abstract:
The cyclooxygenase-2 is a pro-inflammatory and cancer marker, whose mRNA stability and translation is regulated by the CUG-binding protein 2 interacting with AU-rich sequences in the 3' untranslated region. Here, we present the solution NMR structure of CUG-binding protein 2 RRM3 in complex with 5'-UUUAA-3' originating from the COX-2 3'-UTR. We show that RRM3 uses the same binding surface and protein moieties to interact with AU- and UG-rich RNA motifs, binding with low and high affinity, respectively. Using NMR spectroscopy, isothermal titration calorimetry and molecular dynamics simulations, we demonstrate that distinct sub-states characterized by different aromatic side-chain conformations at the RNA-binding surface allow for high- or low-affinity binding with functional implications. This study highlights a mechanism for RNA discrimination possibly common to multiple RRMs as several prominent members display a similar rearrangement of aromatic residues upon binding their targets.The RNA Recognition Motif (RRM) is the most ubiquitous RNA binding domain. Here the authors combined NMR and molecular dynamics simulations and show that the RRM RNA binding surface exists in different states and that a conformational switch of aromatic side-chains fine-tunes sequence specific binding affinities.
Organizational Affiliation:
Department of Biology, ETH Zürich, Institute of Molecular Biology and Biophysics, HPP L 14.1 Hönggerbergring 64, 8093, Zürich, Switzerland.