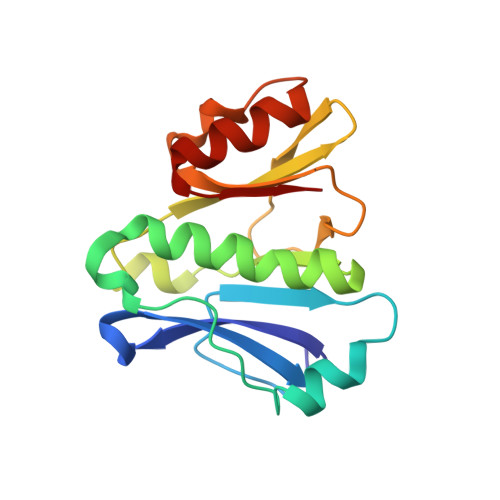
Toxoplasma gondii immune mapped protein 1 is anchored to the inner leaflet of the plasma membrane and adopts a novel protein fold.
Jia, Y., Benjamin, S., Liu, Q., Xu, Y., Dogga, S.K., Liu, J., Matthews, S., Soldati-Favre, D.(2016) Biochim Biophys Acta 1865: 208-219
- PubMed: 27888074
- DOI: https://doi.org/10.1016/j.bbapap.2016.11.010
- Primary Citation of Related Structures:
5LG9 - PubMed Abstract:
The immune mapped protein 1 (IMP1) was first identified as a protective antigen in Eimeria maxima and described as vaccine candidate and invasion factor in Toxoplasma gondii. We show here that TgIMP1 localizes to the inner leaflet of plasma membrane (PM) via dual acylation. Mutations either in the N-terminal myristoylation or palmitoylation sites (G2 and C5) cause relocalization of TgIMP1 to the cytosol. The first 11 amino acids are sufficient for PM targeting and the presence of lysine (K7) is critical. Disruption of TgIMP1 gene by double homologous recombination revealed no invasion defect or any measurable alteration in the lytic cycle of tachyzoites. Following immunization with TgIMP1 DNA vaccine, mice challenged with either wild type or IMP1-ko parasites showed no significant difference in protection. The sequence analysis identified a structured C-terminal domain that is present in a broader family of IMP1-like proteins conserved across the members of Apicomplexa. We present the solution structure of this domain determined from NMR data and describe a new protein fold not seen before.
Organizational Affiliation:
Department of Microbiology and Molecular Medicine, CMU, University of Geneva, Rue Michel-Servet 1, CH-1211 Geneva, Switzerland.