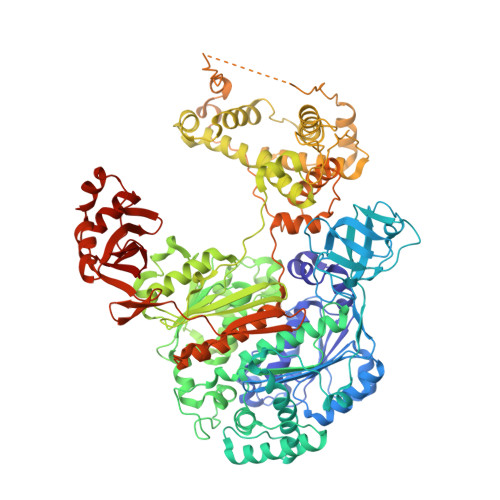
S. pombe Uba1-Ubc15 Structure Reveals a Novel Regulatory Mechanism of Ubiquitin E2 Activity.
Lv, Z., Rickman, K.A., Yuan, L., Williams, K., Selvam, S.P., Woosley, A.N., Howe, P.H., Ogretmen, B., Smogorzewska, A., Olsen, S.K.(2017) Mol Cell 65: 699-714.e6
- PubMed: 28162934
- DOI: https://doi.org/10.1016/j.molcel.2017.01.008
- Primary Citation of Related Structures:
5KNL - PubMed Abstract:
Ubiquitin (Ub) E1 initiates the Ub conjugation cascade by activating and transferring Ub to tens of different E2s. How Ub E1 cooperates with E2s that differ substantially in their predicted E1-interacting residues is unknown. Here, we report the structure of S. pombe Uba1 in complex with Ubc15, a Ub E2 with intrinsically low E1-E2 Ub thioester transfer activity. The structure reveals a distinct Ubc15 binding mode that substantially alters the network of interactions at the E1-E2 interface compared to the only other available Ub E1-E2 structure. Structure-function analysis reveals that the intrinsically low activity of Ubc15 largely results from the presence of an acidic residue at its N-terminal region. Notably, Ub E2 N termini are serine/threonine rich in many other Ub E2s, leading us to hypothesize that phosphorylation of these sites may serve as a novel negative regulatory mechanism of Ub E2 activity, which we demonstrate biochemically and in cell-based assays.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425, USA; Hollings Cancer Center, Medical University of South Carolina, Charleston, SC 29425, USA.