Structural and mechanistic insights into the biosynthesis of CDP-archaeol in membranes.
Ren, S., Caforio, A., Yang, Q., Sun, B., Yu, F., Zhu, X., Wang, J., Dou, C., Fu, Q., Huang, N., Sun, Q., Nie, C., Qi, S., Gong, X., He, J., Wei, Y., Driessen, A.J., Cheng, W.(2017) Cell Res 27: 1378-1391
- PubMed: 28961231
- DOI: https://doi.org/10.1038/cr.2017.122
- Primary Citation of Related Structures:
5GUF - PubMed Abstract:
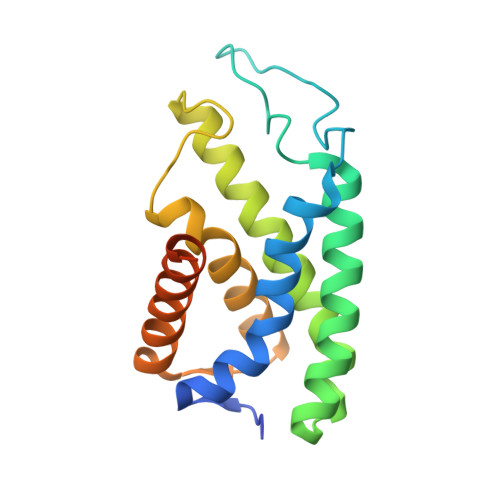
The divergence of archaea, bacteria and eukaryotes was a fundamental step in evolution. One marker of this event is a major difference in membrane lipid chemistry between these kingdoms. Whereas the membranes of bacteria and eukaryotes primarily consist of straight fatty acids ester-bonded to glycerol-3-phosphate, archaeal phospholipids consist of isoprenoid chains ether-bonded to glycerol-1-phosphate. Notably, the mechanisms underlying the biosynthesis of these lipids remain elusive. Here, we report the structure of the CDP-archaeol synthase (CarS) of Aeropyrum pernix (ApCarS) in the CTP- and Mg 2+ -bound state at a resolution of 2.4 Å. The enzyme comprises a transmembrane domain with five helices and cytoplasmic loops that together form a large charged cavity providing a binding site for CTP. Identification of the binding location of CTP and Mg 2+ enabled modeling of the specific lipophilic substrate-binding site, which was supported by site-directed mutagenesis, substrate-binding affinity analyses, and enzyme assays. We propose that archaeol binds within two hydrophobic membrane-embedded grooves formed by the flexible transmembrane helix 5 (TM5), together with TM1 and TM4. Collectively, structural comparisons and analyses, combined with functional studies, not only elucidated the mechanism governing the biosynthesis of phospholipids with ether-bonded isoprenoid chains by CTP transferase, but also provided insights into the evolution of this enzyme superfamily from archaea to bacteria and eukaryotes.
Organizational Affiliation:
Division of Respiratory and Critical Care Medicine, State Key Laboratory of Biotherapy, West China Hospital of Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu, Sichuan 610041, China.