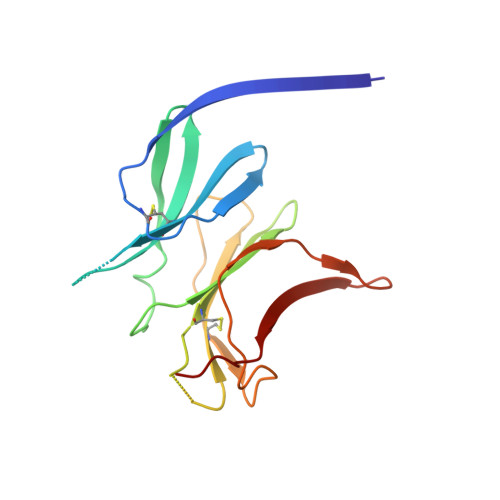
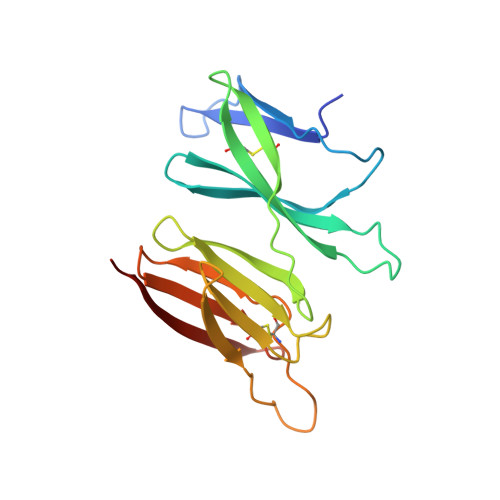
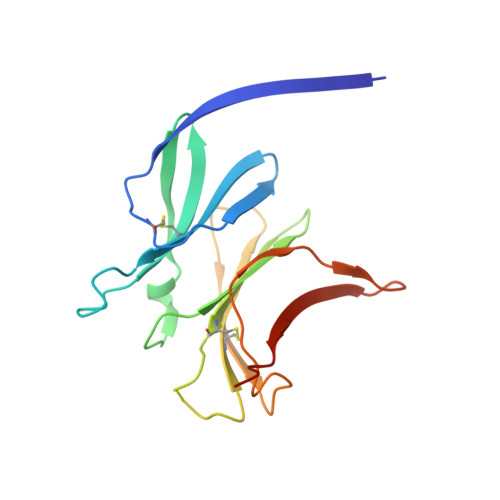
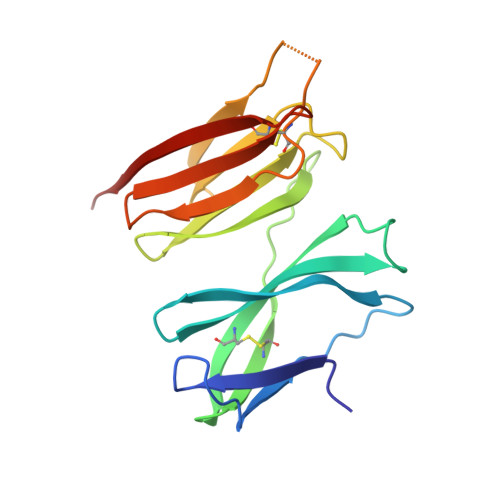
Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Favuzza, P., Guffart, E., Tamborrini, M., Scherer, B., Dreyer, A.M., Rufer, A.C., Erny, J., Hoernschemeyer, J., Thoma, R., Schmid, G., Gsell, B., Lamelas, A., Benz, J., Joseph, C., Matile, H., Pluschke, G., Rudolph, M.G.(2017) Elife 6
- PubMed: 28195038
- DOI: https://doi.org/10.7554/eLife.20383
- Primary Citation of Related Structures:
5EZI, 5EZJ, 5EZL, 5EZN, 5EZO - PubMed Abstract:
Invasion of erythrocytes by Plasmodial merozoites is a composite process involving the interplay of several proteins. Among them, the Plasmodium falciparum Cysteine-Rich Protective Antigen (PfCyRPA) is a crucial component of a ternary complex, including Reticulocyte binding-like Homologous protein 5 (PfRH5) and the RH5-interacting protein (PfRipr), essential for erythrocyte invasion. Here, we present the crystal structures of PfCyRPA and its complex with the antigen-binding fragment of a parasite growth inhibitory antibody. PfCyRPA adopts a 6-bladed β-propeller structure with similarity to the classic sialidase fold, but it has no sialidase activity and fulfills a purely non-enzymatic function. Characterization of the epitope recognized by protective antibodies may facilitate design of peptidomimetics to focus vaccine responses on protective epitopes. Both in vitro and in vivo anti-PfCyRPA and anti-PfRH5 antibodies showed more potent parasite growth inhibitory activity in combination than on their own, supporting a combined delivery of PfCyRPA and PfRH5 in vaccines.
Organizational Affiliation:
Medical Parasitology and Infection Biology Department, Swiss Tropical and Public Health Institute, Basel, Switzerland.