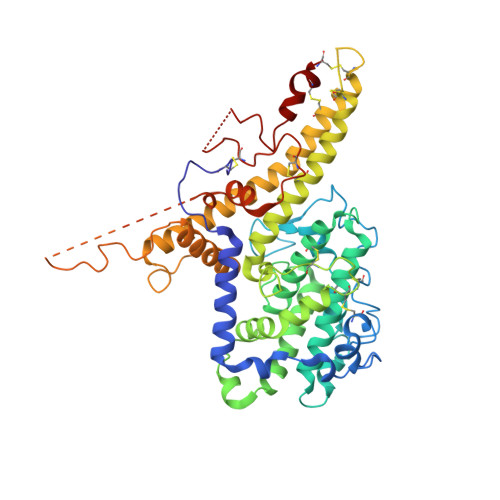
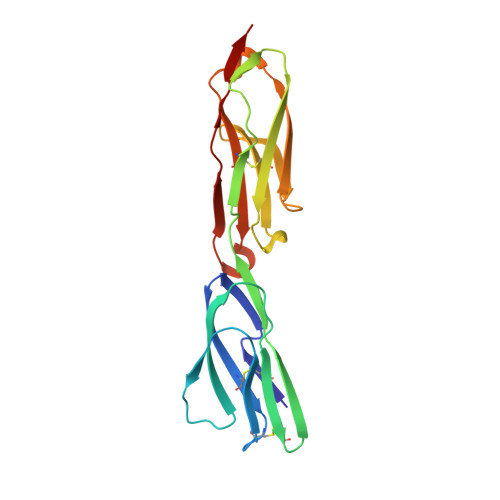
Structure-Guided Identification of a Family of Dual Receptor-Binding PfEMP1 that Is Associated with Cerebral Malaria.
Lennartz, F., Adams, Y., Bengtsson, A., Olsen, R.W., Turner, L., Ndam, N.T., Ecklu-Mensah, G., Moussiliou, A., Ofori, M.F., Gamain, B., Lusingu, J.P., Petersen, J.E., Wang, C.W., Nunes-Silva, S., Jespersen, J.S., Lau, C.K., Theander, T.G., Lavstsen, T., Hviid, L., Higgins, M.K., Jensen, A.T.(2017) Cell Host Microbe 21: 403-414
- PubMed: 28279348
- DOI: https://doi.org/10.1016/j.chom.2017.02.009
- Primary Citation of Related Structures:
5MZA - PubMed Abstract:
Cerebral malaria is a deadly outcome of infection by Plasmodium falciparum, occurring when parasite-infected erythrocytes accumulate in the brain. These erythrocytes display parasite proteins of the PfEMP1 family that bind various endothelial receptors. Despite the importance of cerebral malaria, a binding phenotype linked to its symptoms has not been identified. Here, we used structural biology to determine how a group of PfEMP1 proteins interacts with intercellular adhesion molecule 1 (ICAM-1), allowing us to predict binders from a specific sequence motif alone. Analysis of multiple Plasmodium falciparum genomes showed that ICAM-1-binding PfEMP1s also interact with endothelial protein C receptor (EPCR), allowing infected erythrocytes to synergistically bind both receptors. Expression of these PfEMP1s, predicted to bind both ICAM-1 and EPCR, is associated with increased risk of developing cerebral malaria. This study therefore reveals an important PfEMP1-binding phenotype that could be targeted as part of a strategy to prevent cerebral malaria.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, OX1 3QU Oxford, UK.