Structural Congruency of Ligand Binding to the Insulin and Insulin/Type 1 Insulin-like Growth Factor Hybrid Receptors.
Menting, J.G., Lawrence, C.F., Kong, G.K., Margetts, M.B., Ward, C.W., Lawrence, M.C.(2015) Structure 23: 1271-1282
- PubMed: 26027733
- DOI: https://doi.org/10.1016/j.str.2015.04.016
- Primary Citation of Related Structures:
4XSS, 4XST - PubMed Abstract:
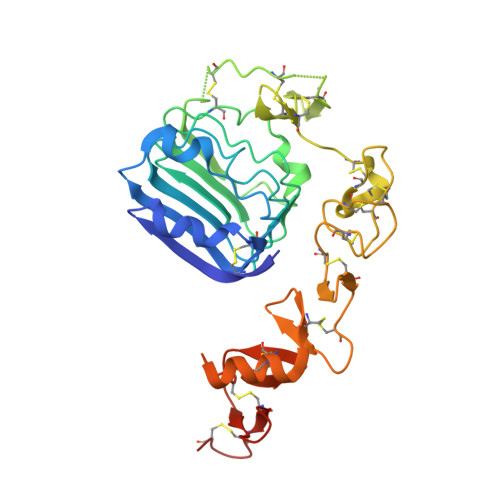
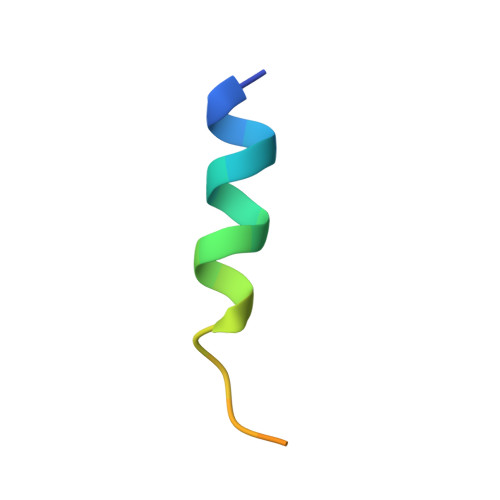
The homodimeric insulin and type 1 insulin-like growth factor receptors (IR and IGF-1R) share a common architecture and each can bind all three ligands within the family: insulin and insulin-like growth factors I and II (IGF-I and IFG-II). The receptor monomers also assemble as heterodimers, the primary ligand-binding sites of which each comprise the first leucine-rich repeat domain (L1) of one receptor type and an α-chain C-terminal segment (αCT) of the second receptor type. We present here crystal structures of IGF-I bound to such a hybrid primary binding site and of a ligand-free version of an IR αCT peptide bound to an IR L1 plus cysteine-rich domain construct (IR310.T). These structures, refined at 3.0-Å resolution, prove congruent to respective existing structures of insulin-complexed IR310.T and the intact apo-IR ectodomain. As such, they provide key missing links in the emerging, but sparse, repertoire of structures defining the receptor family.
Organizational Affiliation:
Structural Biology Division, Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia.