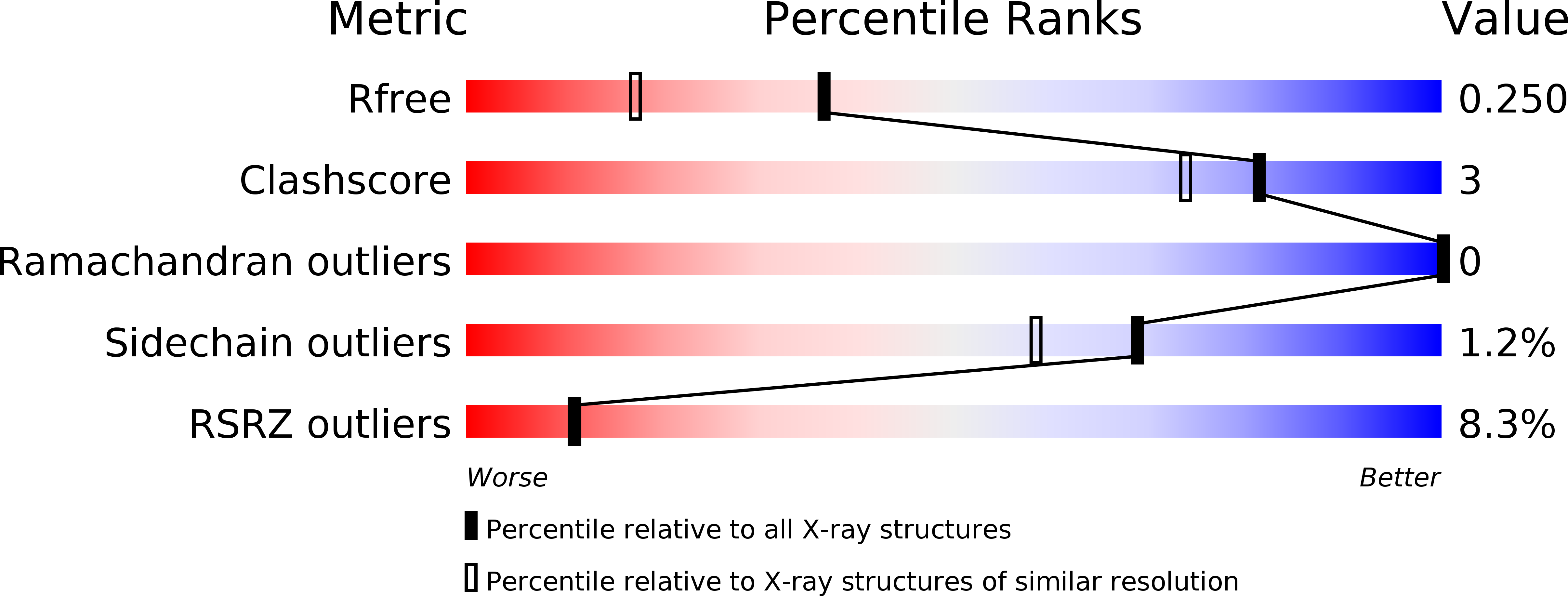
VP22 core domain from Herpes simplex virus 1 reveals a surprising structural conservation in both the Alpha- and Gammaherpesvirinae subfamilies.
Hew, K., Dahlroth, S.L., Pan, L.X., Cornvik, T., Nordlund, P.(2015) J Gen Virol 96: 1436-1445
- PubMed: 26068188
- DOI: https://doi.org/10.1099/vir.0.000078
- Primary Citation of Related Structures:
4XAL - PubMed Abstract:
The viral tegument is a layer of proteins between the herpesvirus capsid and its outer envelope. According to phylogenetic studies, only a third of these proteins are conserved amongst the three subfamilies (Alpha-, Beta- and Gammaherpesvirinae) of the family Herpesviridae. Although some of these tegument proteins have been studied in more detail, the structure and function of the majority of them are still poorly characterized. VP22 from Herpes simplex virus 1 (subfamily Alphaherpesvirinae) is a highly interacting tegument protein that has been associated with tegument assembly. We have determined the crystal structure of the conserved core domain of VP22, which reveals an elongated dimer with several potential protein-protein interaction regions and a peptide-binding site. The structure provides us with the structural basics to understand the numerous functional mutagenesis studies of VP22 found in the literature. It also establishes an unexpected structural homology to the tegument protein ORF52 from Murid herpesvirus 68 (subfamily Gammaherpesvirinae). Homologues for both VP22 and ORF52 have been identified in their respective subfamilies. Although there is no obvious sequence overlap in the two subfamilies, this structural conservation provides compelling structural evidence for shared ancestry and functional conservation.
Organizational Affiliation:
Division of Structural Biology and Biochemistry, School of Biological Sciences, Nanyang Technological University, 138673, Singapore.