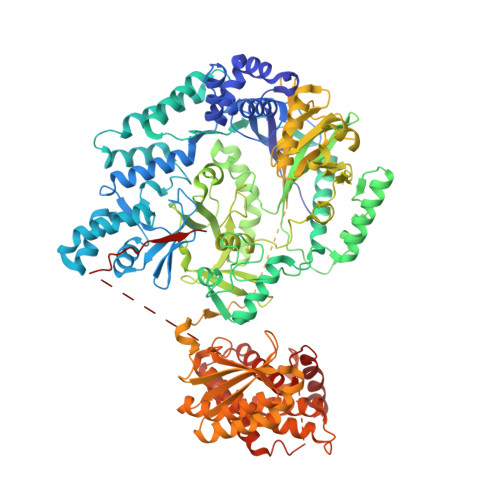
Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB.
Ortega, M.A., Hao, Y., Zhang, Q., Walker, M.C., van der Donk, W.A., Nair, S.K.(2015) Nature 517: 509-512
- PubMed: 25363770
- DOI: https://doi.org/10.1038/nature13888
- Primary Citation of Related Structures:
4WD9 - PubMed Abstract:
Lantibiotics are a class of peptide antibiotics that contain one or more thioether bonds. The lantibiotic nisin is an antimicrobial peptide that is widely used as a food preservative to combat food-borne pathogens. Nisin contains dehydroalanine and dehydrobutyrine residues that are formed by the dehydration of Ser/Thr by the lantibiotic dehydratase NisB (ref. 2). Recent biochemical studies revealed that NisB glutamylates Ser/Thr side chains as part of the dehydration process. However, the molecular mechanism by which NisB uses glutamate to catalyse dehydration remains unresolved. Here we show that this process involves glutamyl-tRNA(Glu) to activate Ser/Thr residues. In addition, the 2.9-Å crystal structure of NisB in complex with its substrate peptide NisA reveals the presence of two separate domains that catalyse the Ser/Thr glutamylation and glutamate elimination steps. The co-crystal structure also provides insights into substrate recognition by lantibiotic dehydratases. Our findings demonstrate an unexpected role for aminoacyl-tRNA in the formation of dehydroamino acids in lantibiotics, and serve as a basis for the functional characterization of the many lantibiotic-like dehydratases involved in the biosynthesis of other classes of natural products.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, USA.