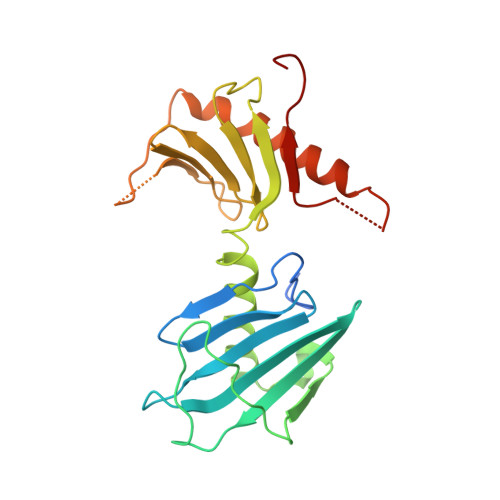
Structure of the C. elegans ZYG-1 Cryptic Polo Box Suggests a Conserved Mechanism for Centriolar Docking of Plk4 Kinases.
Shimanovskaya, E., Viscardi, V., Lesigang, J., Lettman, M.M., Qiao, R., Svergun, D.I., Round, A., Oegema, K., Dong, G.(2014) Structure 22: 1090-1104
- PubMed: 24980795
- DOI: https://doi.org/10.1016/j.str.2014.05.009
- Primary Citation of Related Structures:
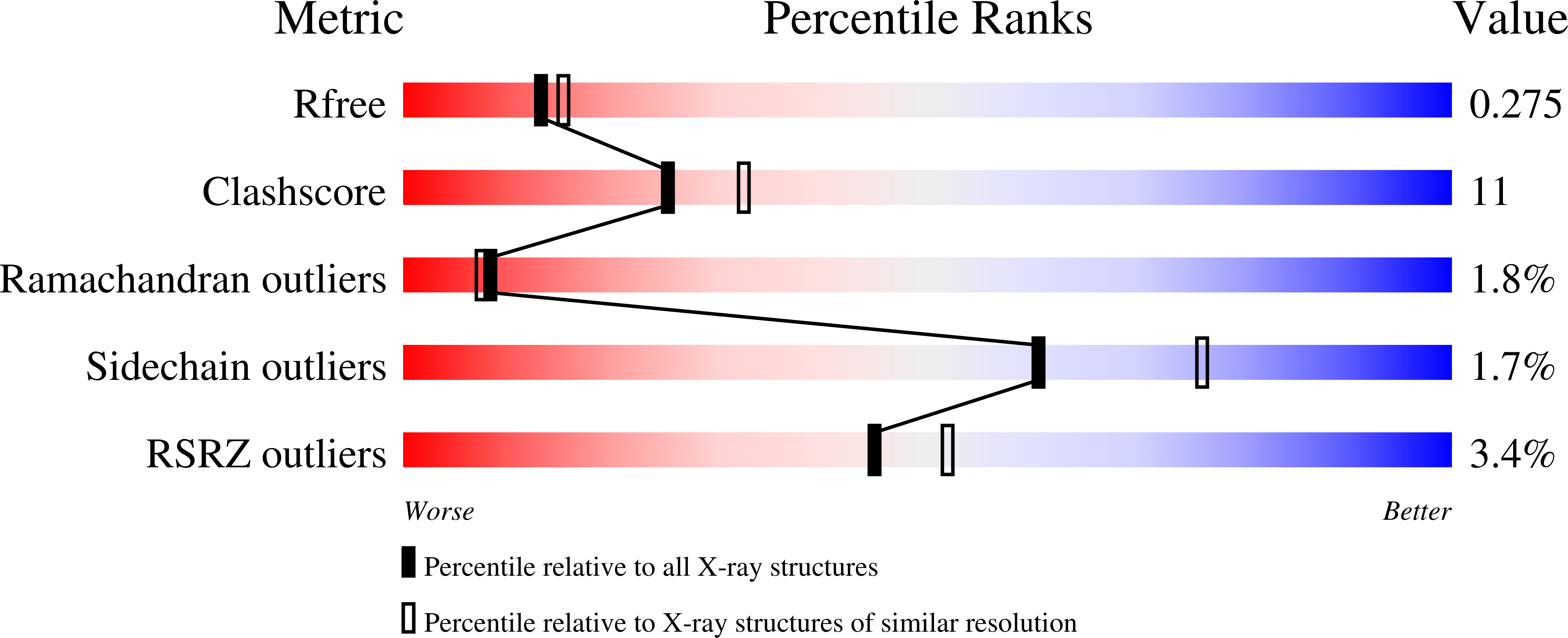
4NK7, 4NKB - PubMed Abstract:
Plk4 family kinases control centriole assembly. Plk4s target mother centrioles through an interaction between their cryptic polo box (CPB) and acidic regions in the centriolar receptors SPD-2/Cep192 and/or Asterless/Cep152. Here, we report a crystal structure for the CPB of C. elegans ZYG-1, which forms a Z-shaped dimer containing an intermolecular β sheet with an extended basic surface patch. Biochemical and in vivo analysis revealed that electrostatic interactions dock the ZYG-1 CPB basic patch onto the SPD-2-derived acidic region to promote ZYG-1 targeting and new centriole assembly. Analysis of a different crystal form of the Drosophila Plk4 (DmPlk4) CPB suggests that it also forms a Z-shaped dimer. Comparison of the ZYG-1 and DmPlk4 CPBs revealed structural changes in the ZYG-1 CPB that confer selectivity for binding SPD-2 over Asterless-derived acidic regions. Overall, our findings suggest a conserved mechanism for centriolar docking of Plk4 homologs that initiate daughter centriole assembly.
Organizational Affiliation:
Max F. Perutz Laboratories, Medical University of Vienna, 1030 Vienna, Austria.