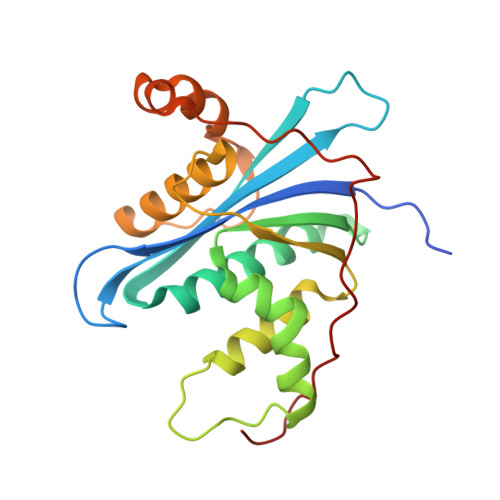
Crystal structure of metagenome-derived LC9-RNase H1 with atypical DEDN active site motif
Nguyen, T.-N., You, D.-J., Kanaya, E., Koga, Y., Kanaya, S.(2013) FEBS Lett 587: 1418-1423
- PubMed: 23523920
- DOI: https://doi.org/10.1016/j.febslet.2013.03.020
- Primary Citation of Related Structures:
4IBN - PubMed Abstract:
The crystal structure of metagenome-derived LC9-RNase H1 was determined. The structure-based mutational analyses indicated that the active site motif of LC9-RNase H1 is altered from DEDD to DEDN. In this motif, the location of the second glutamate residue is moved from αA-helix to β1-strand immediately next to the first aspartate residue, as in the active site of RNase H2. However, the structure and enzymatic properties of LC9-RNase H1 highly resemble those of RNase H1, instead of RNase H2. We propose that LC9-RNase H1 represents bacterial RNases H1 with an atypical DEDN active site motif, which are evolutionarily distinct from those with a typical DEDD active site motif.
Organizational Affiliation:
Department of Material and Life Science, Graduate School of Engineering, Osaka University, 2-1 Yamadaoka, Suita, Osaka 565-0871, Japan.