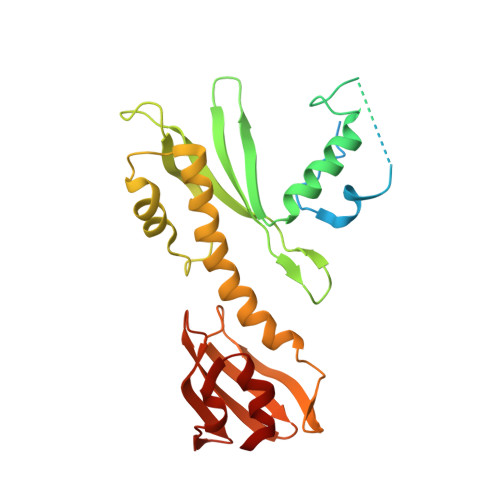
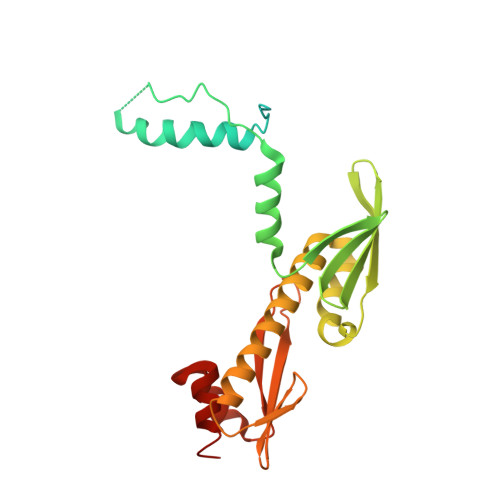
Rwd Domain: A Recurring Module in Kinetochore Architecture Shown by a Ctf19-Mcm21 Complex Structure.
Schmitzberger, F., Harrison, S.C.(2012) EMBO Rep 13: 216
- PubMed: 22322944
- DOI: https://doi.org/10.1038/embor.2012.1
- Primary Citation of Related Structures:
3ZXU - PubMed Abstract:
The proteins Ctf19, Okp1, Mcm21 and Ame1 are the components of COMA, a subassembly of budding-yeast kinetochores. We have determined the crystal structure of a conserved COMA subcomplex--the Ctf19-Mcm21 heterodimer--from Kluyveromyces lactis. Both proteins contain 'double-RWD' domains, which together form a Y-shaped framework with flexible N-terminal extensions. The kinetochore proteins Csm1, Spc24 and Spc25 have related single RWD domains, and Ctf19 and Mcm21 associate with pseudo-twofold symmetry analogous to that in the Csm1 homodimer and the Spc24-Spc25 heterodimer. The double-RWD domain core of the Ctf19-Mcm21 heterodimer is sufficient for association with Okp1-Ame1; the less conserved N-terminal regions may interact with components of a more extensive 'CTF19 complex'. Our structure shows the RWD domain to be a recurring module of kinetochore architecture that may be present in other kinetochore substructures. Like many eukaryotic molecular machines, kinetochores may have evolved from simpler assemblies by multiplication of a few ancestral modules.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 250 Longwood Avenue, Boston, Massachusetts 02115, USA.