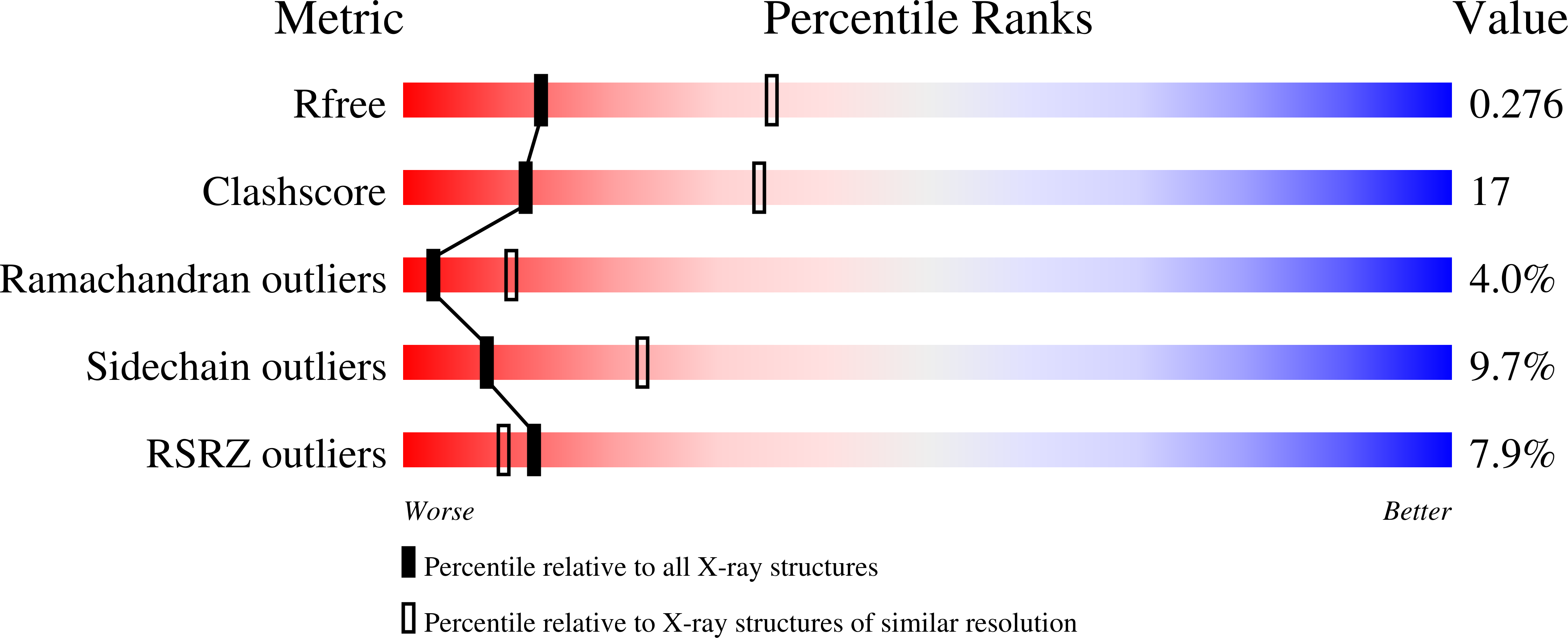
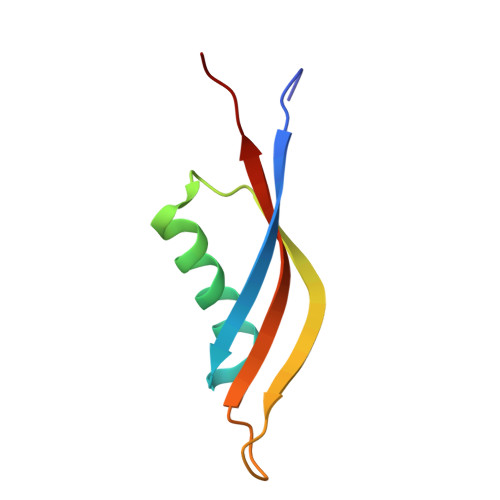
Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
Liu, F., Xiong, J., Kumar, S., Yang, C., Ge, S., Li, S., Xia, N., Swaminathan, K.(2011) J Struct Biol 175: 31-38
- PubMed: 21539921
- DOI: https://doi.org/10.1016/j.jsb.2011.04.013
- Primary Citation of Related Structures:
3OQT - PubMed Abstract:
Dodecins (assembly of twelve monomers) are the smallest known flavoprotein with only 65-73 amino acids and are involved in binding and storage of flavins in archaea. Here we report the crystal structure of Rv1498A, a Mycobacterium tuberculosis dodecin. This bacterial dodecin structure is similar to that of other reported dodecins. Each monomer has a 3 stranded β-sheet and an α-helix perpendicular to it. This protein has polyextreme (halophilic and thermophilic) properties. Interestingly, positively and negatively charged residues aggregate separately and do not seem to contribute to thermophilic and halophilic stability. We have examined the interactions that stabilize the Rv1498A dodecamer by preparing selected point mutants that break salt bridges and hydrophobic contacts, thereby leading to collapse of the assembly.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.