Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Bell, C.H., Porter, S.L., Strawson, A., Stuart, D.I., Armitage, J.P.(2010) PLoS Biol 8: e1000306-e1000306
- PubMed: 20161720
- DOI: https://doi.org/10.1371/journal.pbio.1000306
- Primary Citation of Related Structures:
3KYI, 3KYJ - PubMed Abstract:
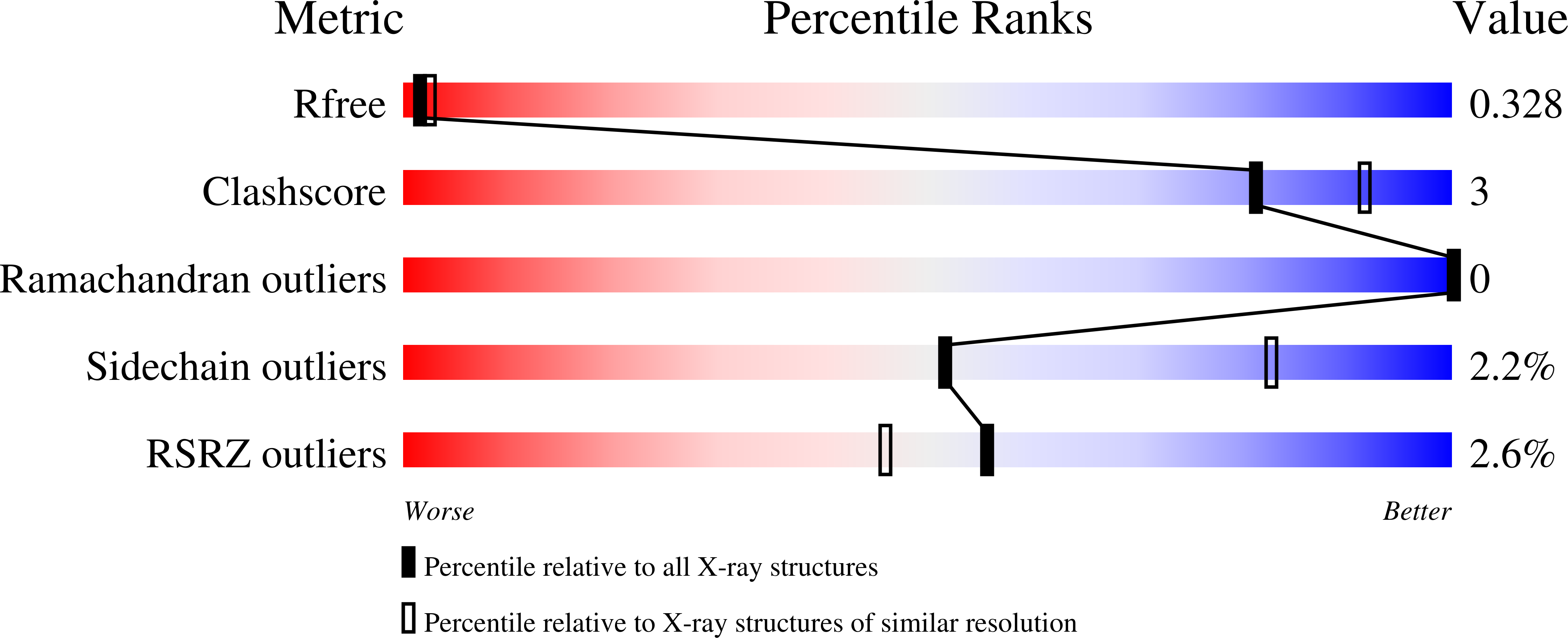
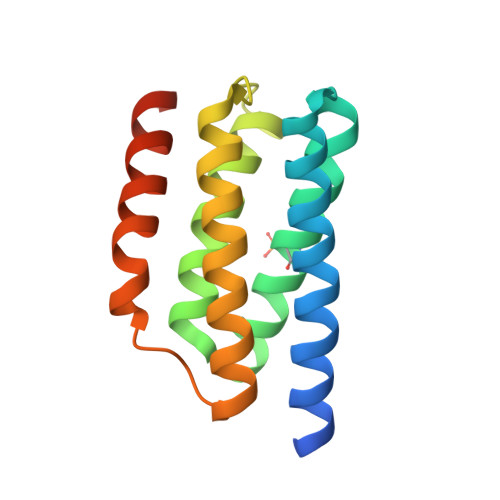
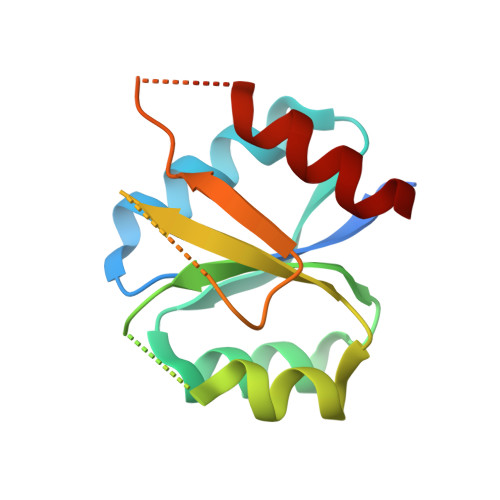
Two-component signal transduction pathways comprising histidine protein kinases (HPKs) and their response regulators (RRs) are widely used to control bacterial responses to environmental challenges. Some bacteria have over 150 different two-component pathways, and the specificity of the phosphotransfer reactions within these systems is tightly controlled to prevent unwanted crosstalk. One of the best understood two-component signalling pathways is the chemotaxis pathway. Here, we present the 1.40 A crystal structure of the histidine-containing phosphotransfer domain of the chemotaxis HPK, CheA(3), in complex with its cognate RR, CheY(6). A methionine finger on CheY(6) that nestles in a hydrophobic pocket in CheA(3) was shown to be important for the interaction and was found to only occur in the cognate RRs of CheA(3), CheY(6), and CheB(2). Site-directed mutagenesis of this methionine in combination with two adjacent residues abolished binding, as shown by surface plasmon resonance studies, and phosphotransfer from CheA(3)-P to CheY(6). Introduction of this methionine and an adjacent alanine residue into a range of noncognate CheYs, dramatically changed their specificity, allowing protein interaction and rapid phosphotransfer from CheA(3)-P. The structure presented here has allowed us to identify specificity determinants for the CheA-CheY interaction and subsequently to successfully reengineer phosphotransfer signalling. In summary, our results provide valuable insight into how cells mediate specificity in one of the most abundant signalling pathways in biology, two-component signal transduction.
Organizational Affiliation:
Oxford Centre for Integrative Systems Biology, Department of Biochemistry, University of Oxford, Oxford, United Kingdom.