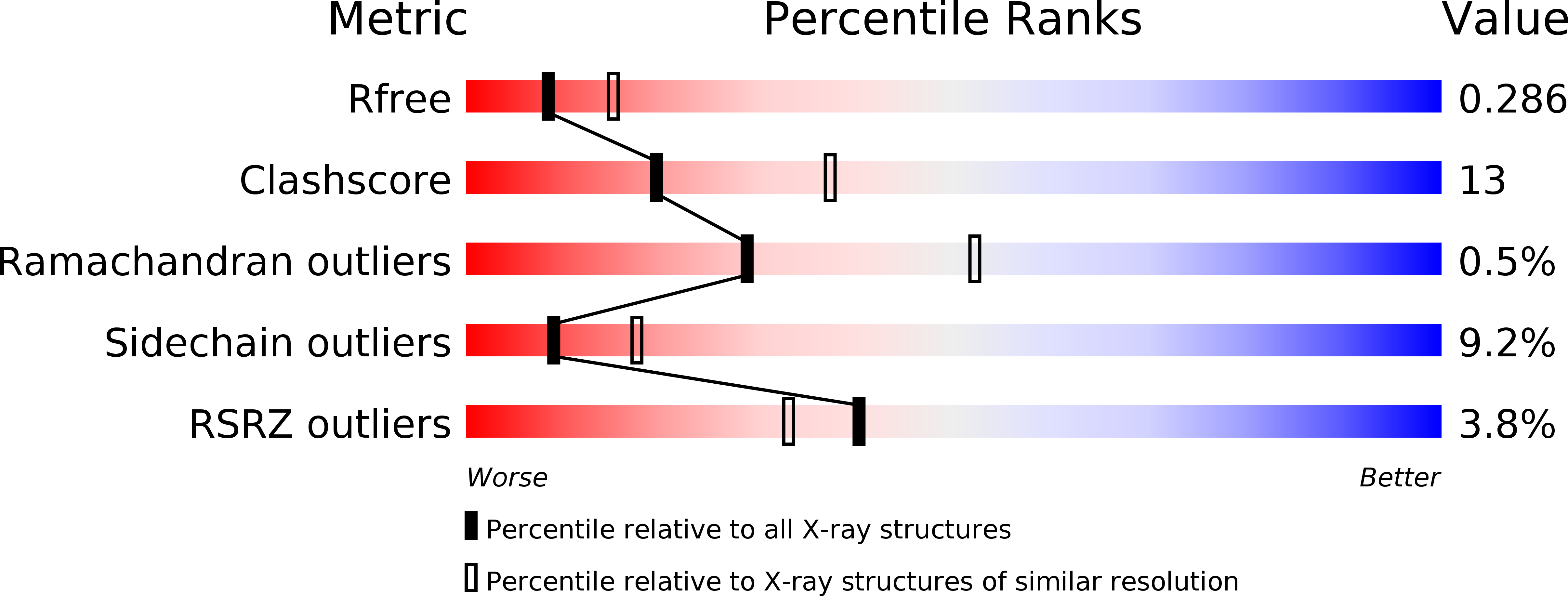
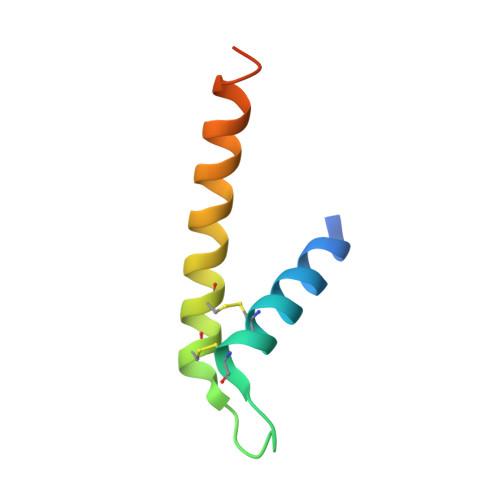
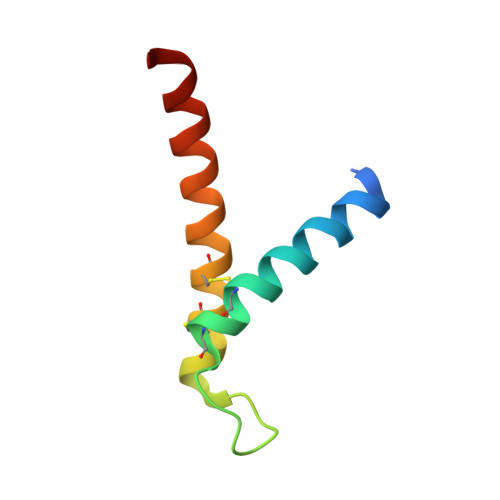
The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23
Beverly, K.N., Sawaya, M.R., Schmid, E., Koehler, C.M.(2008) J Mol Biol 382: 1144-1156
- PubMed: 18706423
- DOI: https://doi.org/10.1016/j.jmb.2008.07.069
- Primary Citation of Related Structures:
3CJH - PubMed Abstract:
The Tim8-Tim13 complex, located in the mitochondrial intermembrane space, functions in the TIM22 import pathway that mediates the import of the mitochondrial carriers Tim23, Tim22, and Tim17 into the mitochondrial inner membrane. The Tim8-Tim13 complex assembles as a hexamer and binds to the substrate Tim23 to chaperone the hydrophobic Tim23 across the aqueous intermembrane space. However, both structural features of the Tim8-Tim13 complex and the binding interaction to Tim23 remain poorly defined. The crystal structure of the yeast Tim8-Tim13 complex, reported here at 2.6 A resolution, reveals that the architecture of the Tim8-Tim13 complex is similar to those of other chaperones such as Tim9-Tim10, prefoldin, and Skp, in which long helices extend from a central body like tentacles from a jellyfish. Surface plasmon resonance was applied to investigate interactions between the Tim8-Tim13 complex and Tim23. The Tim8-Tim13 complex contained approximately six binding sites and showed a complex binding interaction indicative of positive cooperativity rather than a simple bimolecular interaction. By combining results from the structural and binding studies, we provide a molecular model of the Tim8-Tim13 complex binding to Tim23. The regions where the tentacle helices attach to the body of the Tim8-Tim13 complex contain six hydrophobic pockets that likely interact with specific sequences of Tim23 and possibly other substrates. Smaller hydrophobic patches on the tentacles themselves likely interact nonspecifically with the substrate's transmembrane helices, shielding it from the aqueous intermembrane space. The central region of Tim23, which enters the intermembrane space first, may serve to nucleate the binding of the Tim8-Tim13 complex, thereby initiating the chaperoned translocation of Tim23 to the mitochondrial inner membrane.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Box 951569, University of California, Los Angeles, CA 90095-1569, USA.