The C-Terminal Region of Ge-1 Presents Conserved Structural Features Required for P-Body Localization.
Jinek, M., Eulalio, A., Lingel, A., Helms, S., Conti, E., Izaurralde, E.(2008) RNA 14: 1991
- PubMed: 18755833
- DOI: https://doi.org/10.1261/rna.1222908
- Primary Citation of Related Structures:
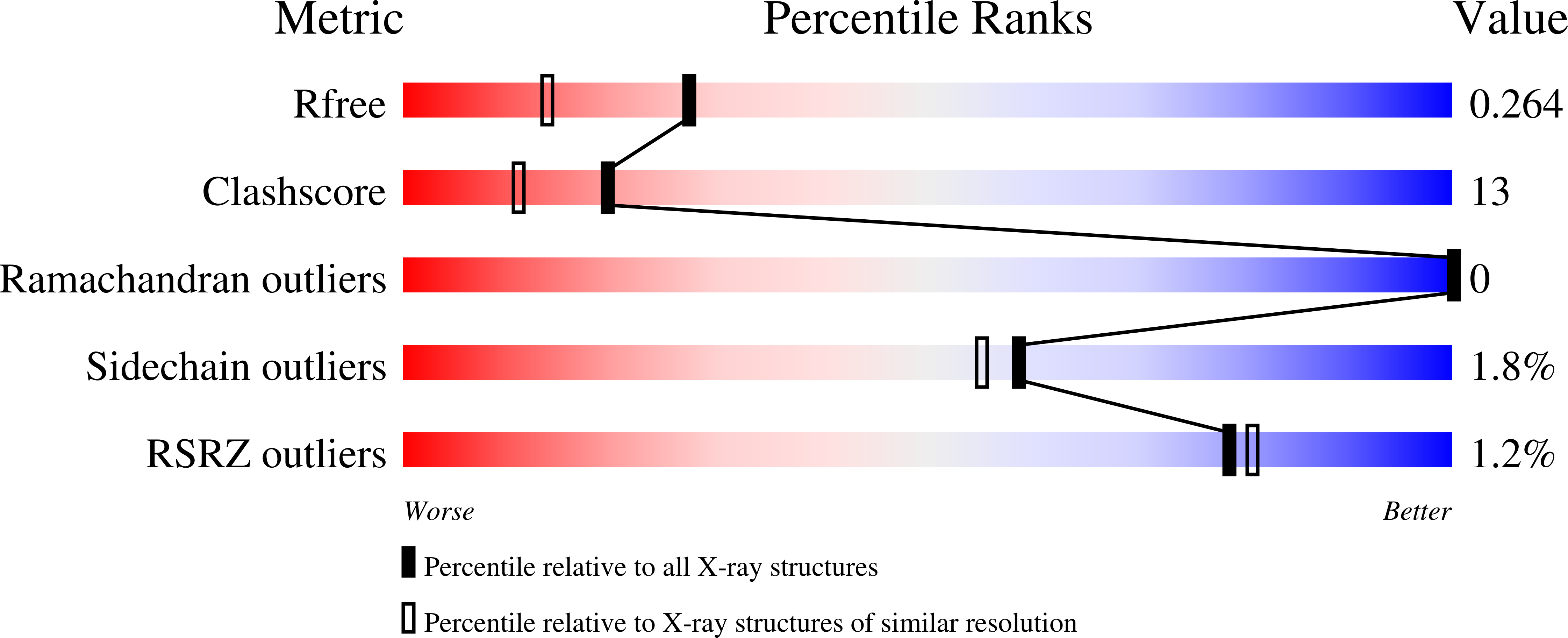
2VXG - PubMed Abstract:
The removal of the 5' cap structure by the DCP1-DCP2 decapping complex irreversibly commits eukaryotic mRNAs to degradation. In human cells, the interaction between DCP1 and DCP2 is bridged by the Ge-1 protein. Ge-1 contains an N-terminal WD40-repeat domain connected by a low-complexity region to a conserved C-terminal domain. It was reported that the C-terminal domain interacts with DCP2 and mediates Ge-1 oligomerization and P-body localization. To understand the molecular basis for these functions, we determined the three-dimensional crystal structure of the most conserved region of the Drosophila melanogaster Ge-1 C-terminal domain. The region adopts an all alpha-helical fold related to ARM- and HEAT-repeat proteins. Using structure-based mutants we identified an invariant surface residue affecting P-body localization. The conservation of critical surface and structural residues suggests that the C-terminal region adopts a similar fold with conserved functions in all members of the Ge-1 protein family.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, D-82152 Martinsried, Germany.