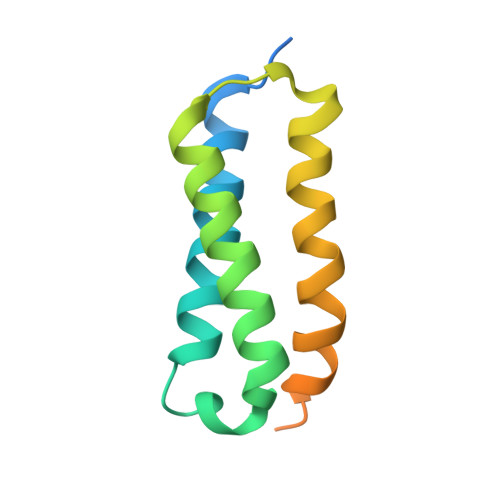
Crystal structure of the C-terminal three-helix bundle subdomain of C. elegans Hsp70.
Worrall, L.J., Walkinshaw, M.D.(2007) Biochem Biophys Res Commun 357: 105-110
- PubMed: 17407764
- DOI: https://doi.org/10.1016/j.bbrc.2007.03.107
- Primary Citation of Related Structures:
2P32 - PubMed Abstract:
Hsp70 chaperones are composed of two domains; the 40 kDa N-terminal nucleotide-binding domain (NDB) and the 30 kDa C-terminal substrate-binding domain (SBD). Structures of the SBD from Escherichia coli homologues DnaK and HscA show it can be further divided into an 18 kDa beta-sandwich subdomain, which forms the hydrophobic binding pocket, and a 10 kDa C-terminal three-helix bundle that forms a lid over the binding pocket. Across prokaryotes and eukaryotes, the NBD and beta-sandwich subdomain are well conserved in both sequence and structure. The C-terminal subdomain is, however, more evolutionary variable and the only eukaryotic structure from rat Hsc70 revealed a diverged helix-loop-helix fold. We have solved the crystal structure of the C-terminal 10 kDa subdomain from Caenorhabditis elegans Hsp70 which forms a helical-bundle similar to the prokaryotic homologues. This provides the first confirmation of the structural conservation of this subdomain in eukaryotes. Comparison with the rat structure reveals a domain-swap dimerisation mechanism; however, the C. elegans subdomain exists exclusively as a monomer in solution in agreement with the hypothesis that regions out with the C-terminal subdomain are necessary for Hsp70 self-association.
Organizational Affiliation:
Centre for Translational and Chemical Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JR, UK.