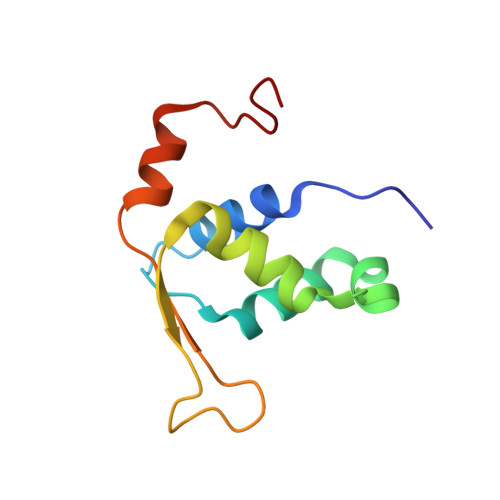
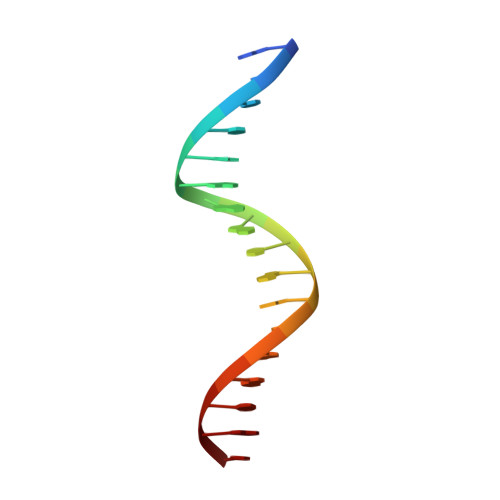
Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex.
Jin, C., Marsden, I., Chen, X., Liao, X.(1999) J Mol Biol 289: 683-690
- PubMed: 10369754
- DOI: https://doi.org/10.1006/jmbi.1999.2819
- Primary Citation of Related Structures:
2HDC - PubMed Abstract:
Genesis is an HNF-3/fkh homologous protein. By using multi-dimensional NMR techniques, we have obtained the solution structure and backbone dynamics of Genesis complexed with a 17 base-pair DNA. Our results indicate that both the local folding and dynamic properties of Genesis are perturbed when it binds to the DNA site. Our data show that a conserved flexible amino acid sequence (wing 1) makes dynamic contacts to DNA in the complex and a short helix is induced by Genesis-DNA interactions. Our data indicate that, unlike the HNF-3gamma/DNA complex, a magnesium ion is not required in forming the stable Genesis-DNA complex.
Organizational Affiliation:
Department or Biochemistry and Molecular Biology, College of Medicine, University of Illinois at Chicago, Chicago, IL, 60612, USA.