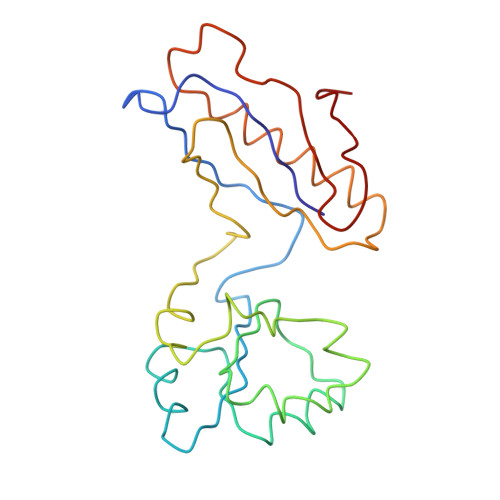
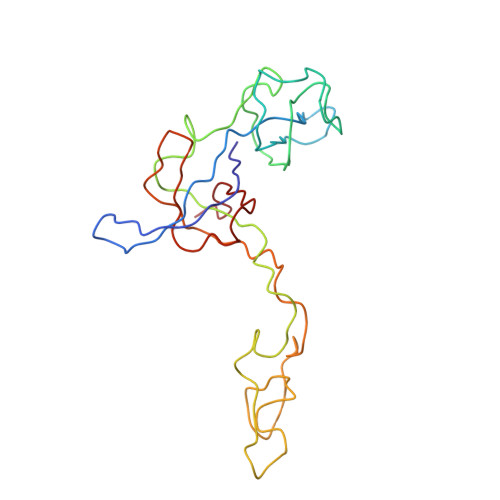
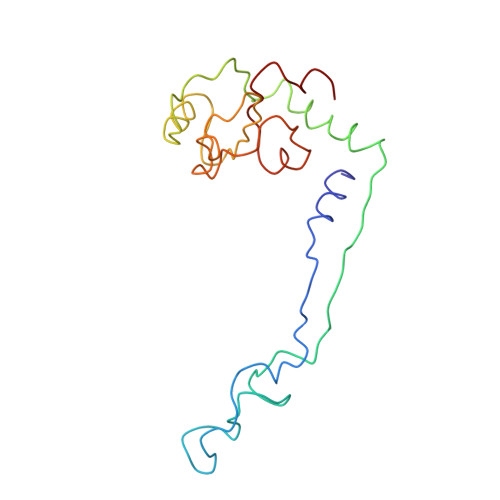
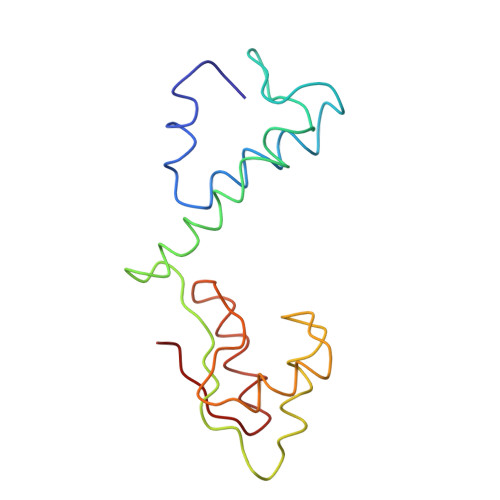
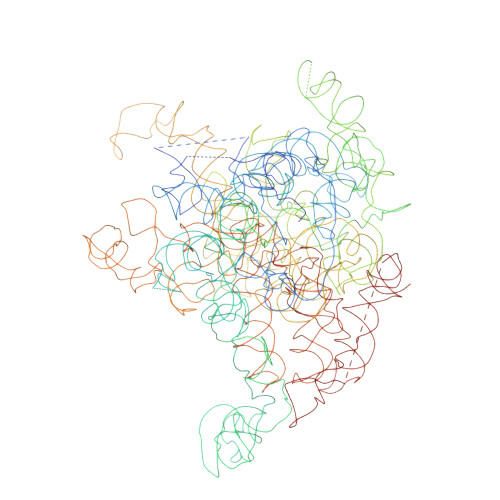
A Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome
Mears, J.A., Sharma, M.R., Gutell, R.R., McCook, A.S., Richardson, P.E., Caulfield, T.R., Agrawal, R.K., Harvey, S.C.(2006) J Mol Biol 358: 193-212
- PubMed: 16510155
- DOI: https://doi.org/10.1016/j.jmb.2006.01.094
- Primary Citation of Related Structures:
2FTC - PubMed Abstract:
Protein translation is essential for all forms of life and is conducted by a macromolecular complex, the ribosome. Evolutionary changes in protein and RNA sequences can affect the 3D organization of structural features in ribosomes in different species. The most dramatic changes occur in animal mitochondria, whose genomes have been reduced and altered significantly. The RNA component of the mitochondrial ribosome (mitoribosome) is reduced in size, with a compensatory increase in protein content. Until recently, it was unclear how these changes affect the 3D structure of the mitoribosome. Here, we present a structural model of the large subunit of the mammalian mitoribosome developed by combining molecular modeling techniques with cryo-electron microscopic data at 12.1A resolution. The model contains 93% of the mitochondrial rRNA sequence and 16 mitochondrial ribosomal proteins in the large subunit of the mitoribosome. Despite the smaller mitochondrial rRNA, the spatial positions of RNA domains known to be involved directly in protein synthesis are essentially the same as in bacterial and archaeal ribosomes. However, the dramatic reduction in rRNA content necessitates evolution of unique structural features to maintain connectivity between RNA domains. The smaller rRNA sequence also limits the likelihood of tRNA binding at the E-site of the mitoribosome, and correlates with the reduced size of D-loops and T-loops in some animal mitochondrial tRNAs, suggesting co-evolution of mitochondrial rRNA and tRNA structures.
Organizational Affiliation:
Department of Biology, Georgia Institute of Technology, Atlanta, GA 30332, USA.