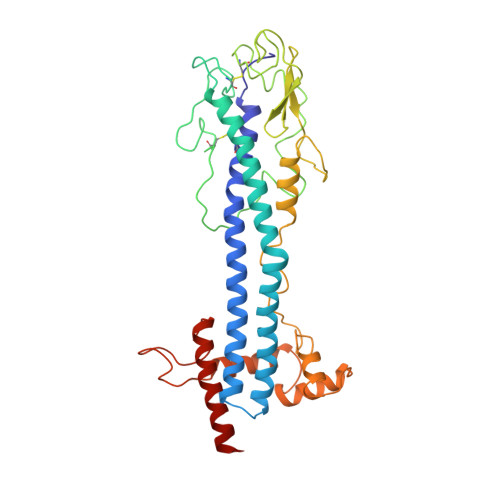
2.9 A resolution structure of the N-terminal domain of a variant surface glycoprotein from Trypanosoma brucei.
Freymann, D., Down, J., Carrington, M., Roditi, I., Turner, M., Wiley, D.(1990) J Mol Biol 216: 141-160
- PubMed: 2231728
- DOI: https://doi.org/10.1016/S0022-2836(05)80066-X
- Primary Citation of Related Structures:
1VSG - PubMed Abstract:
The variant surface glycoprotein (VSG) of Trypanosoma brucei forms a coat on the surface of the parasite; by the expression of a series of antigenically distinct VSGs in the surface coat the parasite escapes the host immune response. The 2.9 A resolution crystal structure of the N-terminal domain of one variant, MITat 1.2, has been determined. The structure was solved using data collected from two crystal forms. Initially a partial model was built into an electron density map based on multiple isomorphous replacement phases and improved by phase combination methods. Subsequently this model was used to obtain the molecular replacement solution for a second crystal form, providing starting phases which were refined using 2-fold non-crystallographic symmetry averaging. The current model includes 362 residues and has been refined using X-PLOR to an R value of 0.22 for data between 7 and 2.9 A. The molecule is a dimer, approximately 100 A long, having an asymmetrical cross section with maximum dimensions of approximately 40 A x 60 A. Two long, approximately 70 A, alpha-helices from each monomer pack together to form, with several other helices, a core helix bundle that extends nearly the full length of the molecule. The "top" of the protein, which in the surface coat may be exposed to the external environment, is formed from the ends of the two long helices, a short three-stranded beta-sheet, and a strand having irregular conformation that packs above these secondary structure elements. Two conserved disulfide bridges are in this part of the molecule. Several elements of the MITat 1.2 sequence, which contribute to the formation of the helix bundle structure, have been identified. These elements can be found in the sequences of several different VSGs, suggesting that to some extent the VSG structure is conserved in those variants.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, MA 02138.