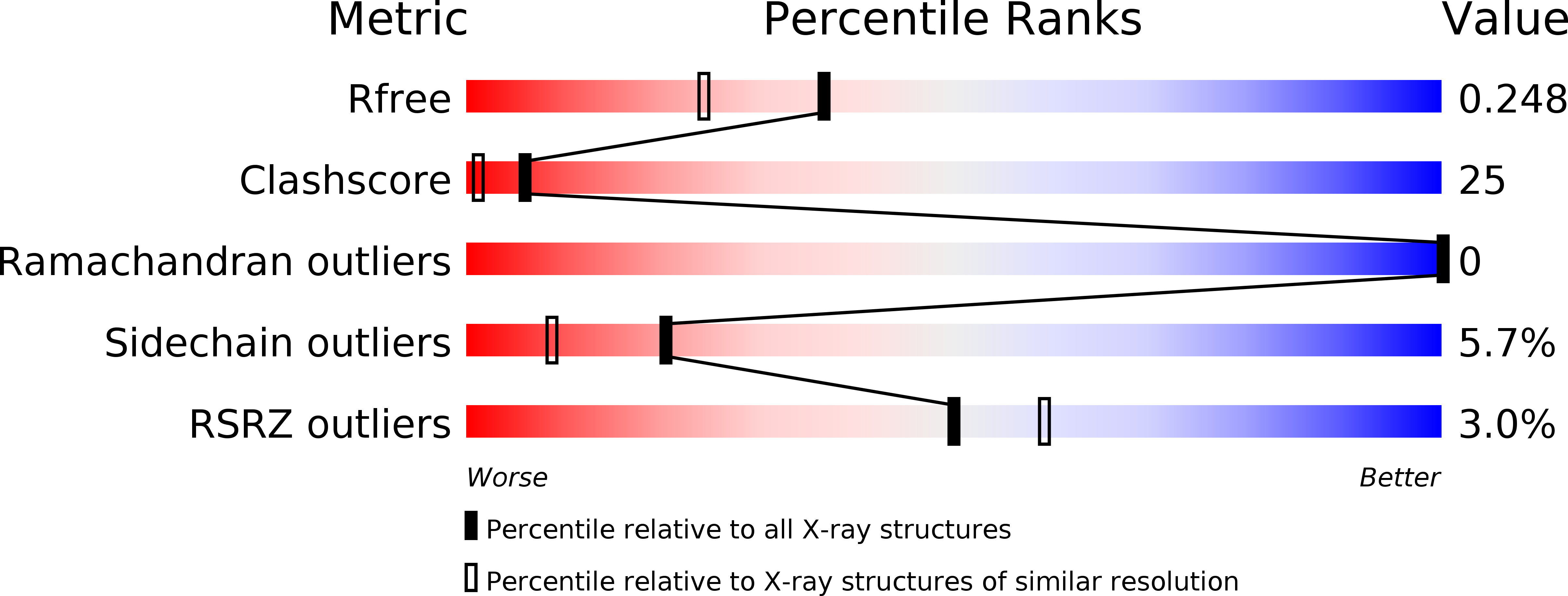
The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor.
Martinez-Hackert, E., Stock, A.M.(1997) Structure 5: 109-124
- PubMed: 9016718
- DOI: https://doi.org/10.1016/s0969-2126(97)00170-6
- Primary Citation of Related Structures:
1OPC - PubMed Abstract:
The differential expression of the ompF and ompC genes is regulated by two proteins that belong to the two component family of signal transduction proteins: the histidine kinase, EnvZ, and the response regulator, OmpR. OmpR belongs to a subfamily of at least 50 response regulators with homologous C-terminal DNA-binding domains of approximately 98 amino acids. Sequence homology with DNA-binding proteins of known structure cannot be detected, and the lack of structural information has prevented understanding of many of this familys functional properties. We have determined the crystal structure of the Escherichia coli OmpR C-terminal domain at 1.95 A resolution. The structure consists of three alpha helices packed against two antiparallel beta sheets. Two helices, alpha2 and alpha3, and the ten residue loop connecting them constitute a variation of the helix-turn-helix (HTH) motif. Helix alpha3 and the loop connecting the two C-terminal beta strands, beta6 and beta7, are probable DNA-recognition sites. Previous mutagenesis studies indicate that the large loop connecting helices alpha2 and alpha3 is the site of interaction with the alpha subunit of RNA polymerase. OmpRc belongs to the family of 'winged helix-turn-helix' DNA-binding proteins. This relationship, and the results from numerous published mutagenesis studies, have helped us to interpret the functions of most of the structural elements present in this protein domain. The structure of OmpRc could be useful in helping to define the positioning of the alpha subunit of RNA polymerase in relation to transcriptional activators that are bound to DNA.
Organizational Affiliation:
Center for Advanced Biotechnology and Medicine, 679 Hoes Lane, Piscataway, NJ 08854, USA.