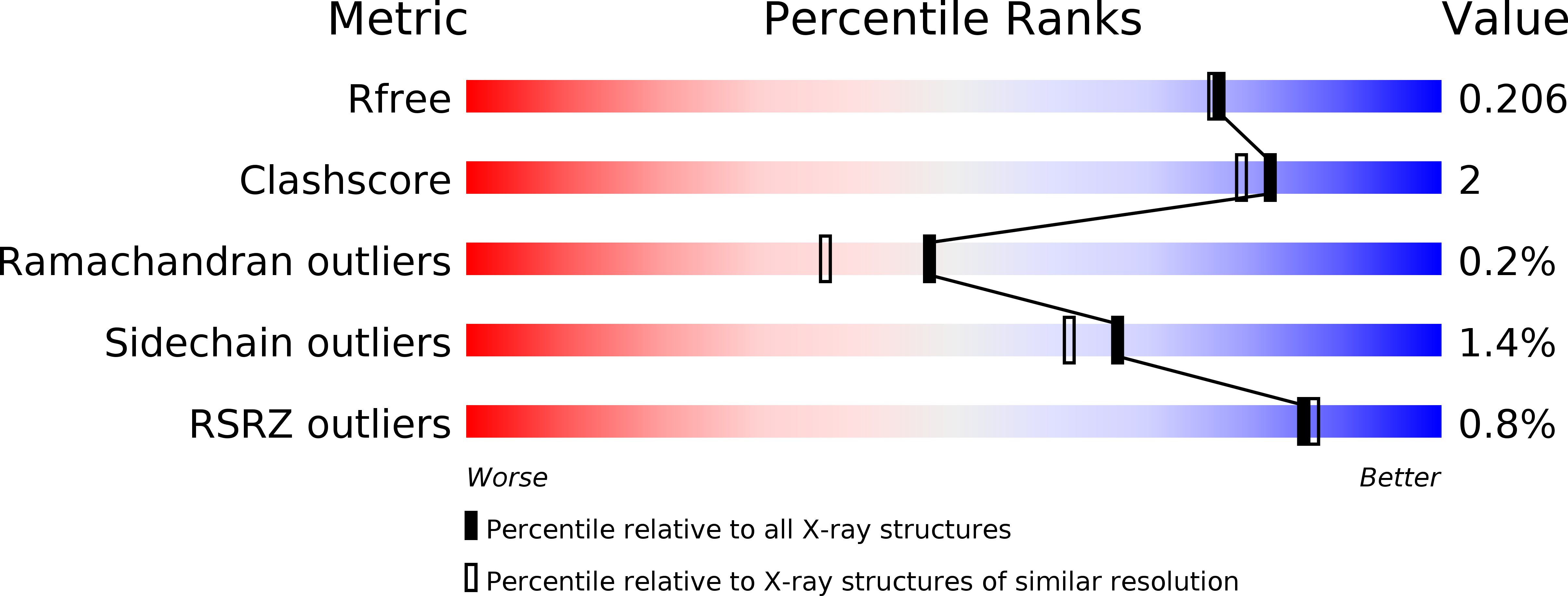
The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
Bashiri, G., Grove, T.L., Hegde, S.S., Lagautriere, T., Gerfen, G.J., Almo, S.C., Squire, C.J., Blanchard, J.S., Baker, E.N.(2019) J Biol Chem 294: 13158-13170
- PubMed: 31315931
- DOI: https://doi.org/10.1074/jbc.RA119.009498
- Primary Citation of Related Structures:
6OVT - PubMed Abstract:
Iron-sulfur clusters are protein cofactors with an ancient evolutionary origin. These clusters are best known for their roles in redox proteins such as ferredoxins, but some iron-sulfur clusters have nonredox roles in the active sites of enzymes. Such clusters are often prone to oxidative degradation, making the enzymes difficult to characterize. Here we report a structural and functional characterization of dihydroxyacid dehydratase (DHAD) from Mycobacterium tuberculosis ( Mtb ), an essential enzyme in the biosynthesis of branched-chain amino acids. Conducting this analysis under fully anaerobic conditions, we solved the DHAD crystal structure, at 1.88 Å resolution, revealing a 2Fe-2S cluster in which one iron ligand is a potentially exchangeable water molecule or hydroxide. UV and EPR spectroscopy both suggested that the substrate binds directly to the cluster or very close to it. Kinetic analysis implicated two ionizable groups in the catalytic mechanism, which we postulate to be Ser-491 and the iron-bound water/hydroxide. Site-directed mutagenesis showed that Ser-491 is essential for activity, and substrate docking indicated that this residue is perfectly placed for proton abstraction. We found that a bound Mg 2+ ion 6.5 Å from the 2Fe-2S cluster plays a key role in substrate binding. We also identified a putative entry channel that enables access to the cluster and show that Mtb -DHAD is inhibited by a recently discovered herbicide, aspterric acid, that, given the essentiality of DHAD for Mtb survival, is a potential lead compound for the design of novel anti-TB drugs.
Organizational Affiliation:
Maurice Wilkins Center for Molecular Biodiscovery and School of Biological Sciences, University of Auckland, Auckland 1010, New Zealand.