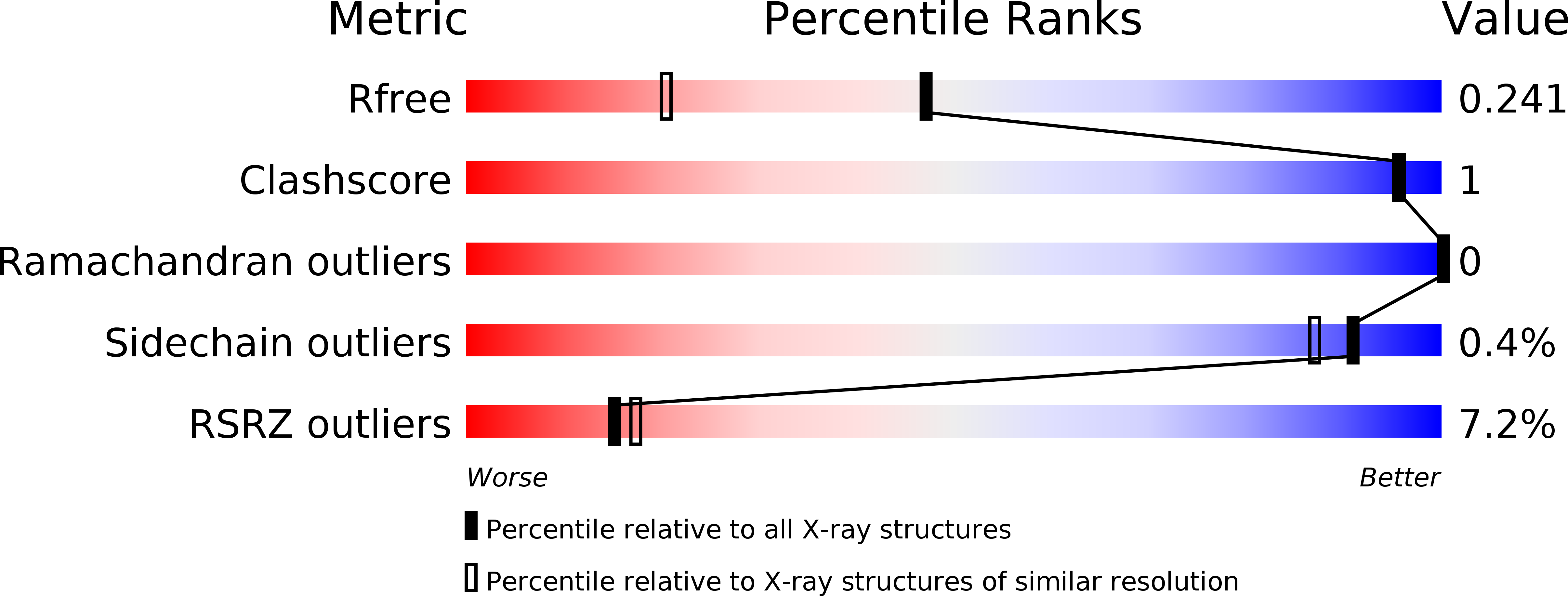
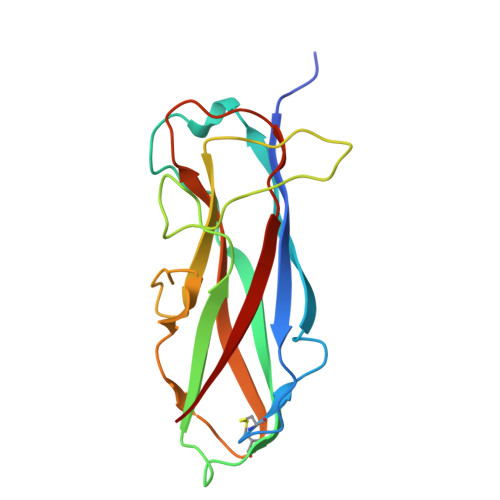
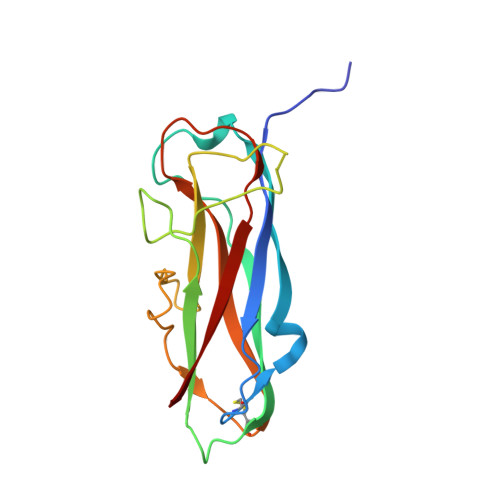
Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
Zyla, D.S., Prota, A.E., Capitani, G., Glockshuber, R.(2019) J Biol Chem 294: 10553-10563
- PubMed: 31126987
- DOI: https://doi.org/10.1074/jbc.RA119.008610
- Primary Citation of Related Structures:
5LP9, 5NKT, 6ERJ - PubMed Abstract:
Adhesive type 1 pili from enteroinvasive, Gram-negative bacteria mediate attachment to host cells. Up to 3000 copies of the main pilus subunit, FimA, assemble into the filamentous, helical quaternary structure of the pilus rod via a mechanism termed donor-strand complementation, in which the N-terminal extension of each subunit, the donor strand, is inserted into the incomplete immunoglobulin-like fold of the preceding FimA subunit. For FimA from Escherichia coli , it has been previously shown that the protein can also adopt a monomeric, self-complemented conformation in which the donor strand is inserted intramolecularly in the opposite orientation relative to that observed for FimA polymers. Notably, soluble FimA monomers can act as apoptosis inhibitors in epithelial cells after uptake of type 1-piliated pathogens. Here, we show that the FimA orthologues from Escherichia coli , Shigella flexneri , and Salmonella enterica can all fold to form self-complemented monomers. We solved X-ray structures of all three FimA monomers at 0.89-1.69 Å resolutions, revealing identical, intramolecular donor-strand complementation mechanisms. Our results also showed that the pseudo-palindromic sequences of the donor strands in all FimA proteins permit their alternative folding possibilities. All FimA monomers proved to be 50-60 kJ/mol less stable against unfolding than their pilus rod-like counterparts (which exhibited very high energy barriers of unfolding and refolding). We conclude that the ability of FimA to adopt an alternative, monomeric state with anti-apoptotic activity is a general feature of FimA proteins of type 1-piliated bacteria.
Organizational Affiliation:
From the Institute of Molecular Biology and Biophysics, ETH Zurich, Otto-Stern-Weg 5, CH-8093 Zurich and.